#new epigenetics tag found
Explore tagged Tumblr posts
Text
1000% not satire, with one (1) caveat: This is very good advice for approaching Cognitive Behavioral Therapy (or is it Dialectal? unfortunately, I'm Ms. Malaprop 2016, from the Jewish-American Princess State, which is probably a politicly incorrect synonym for New York. I thought Serbia was in Syria's location, or maybe vice versa, for most of college, while Serbia was actively at war with one of its neighbors. Oh, and while the state animal or mascot of the J.A.P. State is a highly contested subject, they typical finalist contenders are the Mensch, the Jewish Mother, the Tourist Goy, and the Morbidly Obese Subway Rat.)
Anyhow, where was I? Oh, yes. The psychological phenomenon popularly known as the "placebo effect" or "manifestation" is a very effective component of talk therapy, but please, please don't apply it to personal finance. Get a CFP, or use an online accounting or reliable investment platform. If the money situation is truly dire, it's even pikuach nefash (g-d, did I spell that right?) to use an Intuit product!
Just don't think there's any ethical, anticapitalist way to get rich quick. There isn't. Even the unethical, capitalist ones rarely compile, and if you think otherwise, boy do I have a good deal for you on a piece of the Brooklyn Bridge. I believe in few absolutes, but this one is an exception. Even in a utopian socialist economy, almost everyone who isn't legally exempt has to work to eat.
let go of the old version.
i am right now telling you to let go of the old version of yourself. you are under no obligation to be the person you were yesterday, the person you were some days ago or simply the person you used to be. you are allowed to change. you deserve to change. you can better yourself and become the imagine of yourself you have always wanted to embody.
you don’t need my or anyone’s permission to tell you that you may change to your liking, but sometimes we need to hear it. and that’s why i‘m telling you this!
create a new version.
it’s about damn time you stop pitying yourself and shift your focus back to the basics. you know imagination creates reality. you know that you are in control of your thoughts and you will always be. so, what are you waiting for? you know exactly what to do, so what’s the matter? you are the only person stopping you from reaching your full potential. and i need you to put away all the fears, all of that bottled up anxiety inside of you that prevents you from thinking desirable thoughts. create the version of yourself who you are happy with, the version you want to resemble and do not hold back! write it down, imagine them — how would you act? how would you talk? because whoever that perfect person is that you have in mind, you can be just like them — and NO ONE can stop you from becoming them!
be who you want to be.
i know it can be hard. we often rely on our outer circumstances and follow our outer rules that like to tell us how we need to behave and what we need to do in order to achieve certain things. but that’s irrelevant. all of it — put it AWAY. we already have established that you are god, so STOP living according to these principles. they do not apply to you. they do not apply to GOD.
leave negative feelings behind.
you might be looking for reassurance, for someone to tell you that you can change without feeling bad, humiliated, embarrassed or even ashamed of yourself. changing yourself often means to face the version of you that you no longer desire to identify with. it means to look back to a life that no longer serves you and that you now have to let go of. and you know what? it might be hurtful. but we have been "trained" to be perfect, to do our absolute best, to be okay with the things we are not okay with our whole lives… people have been expecting so much from you — it can be hard to go against those beliefs. but this is manifesting. not being okay with the things you are not okay with and going against them. wanting the best for you. and most importantly, showing yourself the same amount of compassion you show others.
you are allowed to change.
you are allowed to better your family. you are allowed to manifest your love interest. you are allowed to change your past scores. you are allowed to become rich. you are allowed to get whatever it is that you want! this is your reality darling, only yours. i promise, you aren’t hurting anyone BUT yourself if you don’t go for what’s meant for you. because what is life when you can desire but not receive?
⠀⠀⠀⠀⠀⠀⠀⠀⠀⠀⠀⠀⠀⠀⠀ © 23209
#not satire#essay#psychology#psychotherapy#finance and economics#political systems#manifestation#personal finance#get rich quick scams and schemes NOS#placebo effect#new york types slash jewish culture#comedy#longform humor#adhd jokes#every day of my life is an adhd joke: potential memoir title#life advice#because otherwise you get the Holodomor#which means and was roughly the same thing as the Irish Great Hunger#which is also part of my fucked-up genetics#ironically the Holodomor isn't bc the boat got here before WWI#but i bet you can read its handwriting in some of my distant cousins#new epigenetics tag found#epigenetics#social epigenetics#this is too many tags#need a tag-ophage#multidisciplinary polymath#broke polymath problems#DSM 5 or whatever Pokemon generation they're on now#original content slash IP do not Kruppstahl
1K notes
·
View notes
Text
Epigenomic analysis sheds light on risk factors for ALS
New Post has been published on https://sunalei.org/news/epigenomic-analysis-sheds-light-on-risk-factors-for-als/
Epigenomic analysis sheds light on risk factors for ALS
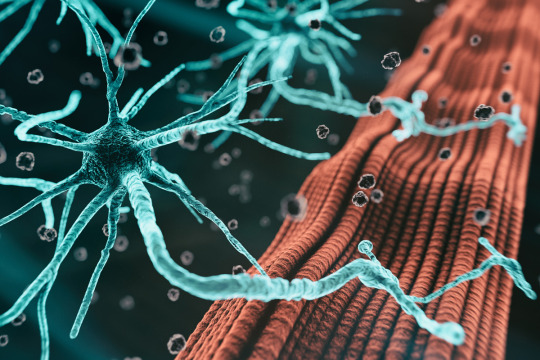
For most patients, it’s unknown exactly what causes amyotrophic lateral sclerosis (ALS), a disease characterized by degeneration of motor neurons that impairs muscle control and eventually leads to death.
Studies have identified certain genes that confer a higher risk of the disease, but scientists believe there are many more genetic risk factors that have yet to be discovered. One reason why these drivers have been hard to find is that some are found in very few patients, making it hard to pick them out without a very large sample of patients. Additionally, some of the risk may be driven by epigenomic factors, rather than mutations in protein-coding genes.
Working with the Answer ALS consortium, a team of MIT researchers has analyzed epigenetic modifications — tags that determine which genes are turned on in a cell — in motor neurons derived from induced pluripotent stem (IPS) cells from 380 ALS patients.
This analysis revealed a strong differential signal associated with a known subtype of ALS, and about 30 locations with modifications that appear to be linked to rates of disease progression in ALS patients. The findings may help scientists develop new treatments that are targeted to patients with certain genetic risk factors.
“If the root causes are different for all these different versions of the disease, the drugs will be very different and the signals in IPS cells will be very different,” says Ernest Fraenkel, the Grover M. Hermann Professor in Health Sciences and Technology in MIT’s Department of Biological Engineering and the senior author of the study. “We may get to a point in a decade or so where we don’t even think of ALS as one disease, where there are drugs that are treating specific types of ALS that only work for one group of patients and not for another.”
MIT postdoc Stanislav Tsitkov is the lead author of the paper, which appears today in Nature Communications.
Finding risk factors
ALS is a rare disease that is estimated to affect about 30,000 people in the United States. One of the challenges in studying the disease is that while genetic variants are believed to account for about 50 percent of ALS risk (with environmental factors making up the rest), most of the variants that contribute to that risk have not been identified.
Similar to Alzheimer’s disease, there may be a large number of genetic variants that can confer risk, but each individual patient may carry only a small number of those. This makes it difficult to identify the risk factors unless scientists have a very large population of patients to analyze.
“Because we expect the disease to be heterogeneous, you need to have large numbers of patients before you can pick up on signals like this. To really be able to classify the subtypes of disease, we’re going to need to look at a lot of people,” Fraenkel says.
About 10 years ago, the Answer ALS consortium began to collect large numbers of patient samples, which could allow for larger-scale studies that might reveal some of the genetic drivers of the disease. From blood samples, researchers can create induced pluripotent stem cells and then induce them to differentiate into motor neurons, the cells most affected by ALS.
“We don’t think all ALS patients are going to be the same, just like all cancers are not the same. And the goal is being able to find drivers of the disease that could be therapeutic targets,” Fraenkel says.
In this study, Fraenkel and his colleagues wanted to see if patient-derived cells could offer any information about molecular differences that are relevant to ALS. They focused on epigenomic modifications, using a method called ATAC-seq to measure chromatin density across the genome of each cell. Chromatin is a complex of DNA and proteins that determines which genes are accessible to be transcribed by the cell, depending on how densely packed the chromatin is.
In data that were collected and analyzed over several years, the researchers did not find any global signal that clearly differentiated the 380 ALS patients in their study from 80 healthy control subjects. However, they did find a strong differential signal associated with a subtype of ALS, characterized by a genetic mutation in the C9orf72 gene.
Additionally, they identified about 30 regions that were associated with slower rates of disease progression in ALS patients. Many of these regions are located near genes related to the cellular inflammatory response; interestingly, several of the identified genes have also been implicated in other neurodegenerative diseases, such as Parkinson’s disease.
“You can use a small number of these epigenomic regions and look at the intensity of the signal there, and predict how quickly someone’s disease will progress. That really validates the hypothesis that the epigenomics can be used as a filter to better understand the contribution of the person’s genome,” Fraenkel says.
“By harnessing the very large number of participant samples and extensive data collected by the Answer ALS Consortium, these studies were able to rigorously test whether the observed changes might be artifacts related to the techniques of sample collection, storage, processing, and analysis, or truly reflective of important biology,” says Lyle Ostrow, an associate professor of neurology at the Lewis Katz School of Medicine at Temple University, who was not involved in the study. “They developed standard ways to control for these variables, to make sure the results can be accurately compared. Such studies are incredibly important for accelerating ALS therapy development, as they will enable data and samples collected from different studies to be analyzed together.”
Targeted drugs
The researchers now hope to further investigate these genomic regions and see how they might drive different aspects of ALS progression in different subsets of patients. This could help scientists develop drugs that might work in different groups of patients, and help them identify which patients should be chosen for clinical trials of those drugs, based on genetic or epigenetic markers.
Last year, the U.S. Food and Drug Administration approved a drug called tofersen, which can be used in ALS patients with a mutation in a gene called SOD1. This drug is very effective for those patients, who make up about 1 percent of the total population of people with ALS. Fraenkel’s hope is that more drugs can be developed for, and tested in, people with other genetic drivers of ALS.
“If you had a drug like tofersen that works for 1 percent of patients and you just gave it to a typical phase two clinical trial, you probably wouldn’t have anybody with that mutation in the trial, and it would’ve failed. And so that drug, which is a lifesaver for people, would never have gotten through,” Fraenkel says.
The MIT team is now using an approach called quantitative trait locus (QTL) analysis to try to identify subgroups of ALS patients whose disease is driven by specific genomic variants.
“We can integrate the genomics, the transcriptomics, and the epigenomics, as a way to find subgroups of ALS patients who have distinct phenotypic signatures from other ALS patients and healthy controls,” Tsitkov says. “We have already found a few potential hits in that direction.”
The research was funded by the Answer ALS program, which is supported by the Robert Packard Center for ALS Research at Johns Hopkins University, Travelers Insurance, ALS Finding a Cure Foundation, Stay Strong Vs. ALS, Answer ALS Foundation, Microsoft, Caterpillar Foundation, American Airlines, Team Gleason, the U.S. National Institutes of Health, Fishman Family Foundation, Aviators Against ALS, AbbVie Foundation, Chan Zuckerberg Initiative, ALS Association, National Football League, F. Prime, M. Armstrong, Bruce Edwards Foundation, the Judith and Jean Pape Adams Charitable Foundation, Muscular Dystrophy Association, Les Turner ALS Foundation, PGA Tour, Gates Ventures, and Bari Lipp Foundation. This work was also supported, in part, by grants from the National Institutes of Health and the MIT-GSK Gertrude B. Elion Research Fellowship Program for Drug Discovery and Disease.
0 notes
Link
0 notes
Text
📆 Aug 2020 📰 'Mono' virus turns on cancer-related genes. Here's how. 🗞 Live Science
A type of herpes virus, one that causes mono, can in rare cases raise the risk of developing certain types of cancer. And now researchers know how: The Epstein-Barr virus (EBV) can directly latch onto bundles of genetic material in infected cells, and switch "on" nearby genes that turn healthy cells cancerous, according to a new study in human cells.
Not all people who become infected with EBV go on to develop cancer; but in rare instances, the virus can raise people's risk of developing nasopharyngeal cancer, Burkitt's lymphoma and certain stomach cancers, according to the American Cancer Society. While more than 90% of people catch the virus worldwide, only about 1.5% of cancer cases are linked to the infection, according to a 2019 report in the journal Annual Review of Pathology. Other viruses that drive cancer growth, such as hepatitis B and human papillomavirus (HPV), do so by worming their way into the genomes of their infected host — but EBV takes a different approach, researchers just found.
Understanding this process could allow scientists to develop drugs and gene therapies to undo the virus's harmful modifications, Rona Scott, an associate professor of microbiology and immunology at Louisiana State University Health Shreveport, who was not involved in the study, told Live Science in an email.
In addition, "identifying footprints [or telltale marks] of EBV infection in cancer may help us determine if EBV, which infects over 95% of adults worldwide, contributes to other cancers not yet associated with this virus," she said.

While some of the details remain fuzzy, "the link between EBV and certain types of cancer has been known for many years," Tan said. For example, the virus has been linked to about 8% to 10% of stomach cancers, which collectively stand as the third leading cause of cancer death globally, according to a statement from Duke-NUS Medical School.
Past research explained one way EBV fuels cancer: The virus triggers chemical reactions that stick molecular tags known as methyl groups onto genes, switching them "on" or "off," according to a 2007 report in the journal Cancer Science. One theory was that these so-called epigenetic modifications, meaning modifications "on top of" the genome, disabled genes that would normally suppress tumor growth.
But Tan wondered whether EBV was also changing the 3D structure of the host genome in ways that up the risk of cancer.
"We were definitely very surprised by the results," as we did not expect the viral genome to directly participate in rewiring the host cell, and controlling which proteins it builds, Tan said.
Even when the authors removed EBV from infected cells, the structural changes the virus made to host DNA stayed put. The finding supports prior evidence that EBV may contribute to cancer in a "hit-and-run" manner, meaning that even if you eliminate the virus itself, the cell's DNA remains altered and continues to drive tumor growth, Scott said.
In addition, "identifying footprints [or telltale marks] of EBV infection in cancer may help us determine if EBV, which infects over 95% of adults worldwide, contributes to other cancers not yet associated with this virus," she said.
0 notes
Text

𝐓𝐀𝐋𝐈𝐀 𝐀𝐘𝐃𝐈𝐍 --- ever heard of curiosity killed the cat ?
( ayca aysin turan / ciswoman / she/her ). introducing talia aydin , the host for the savant. they’ll be twenty - seven years old, and joined the rogues six month ago. you’ll always see a pocket knife around wherever she is.
𝙱𝚄𝚃 𝚆𝙷𝙰𝚃 𝙸𝙵 𝚃𝙷𝙴 𝚆𝙾𝚁𝙻𝙳 𝙸𝚂 𝙰 𝙷𝙾𝙿𝙴𝙻𝙴𝚂𝚂 𝙿𝙻𝙰𝙲𝙴
full name : talia kamile aydin
nickname : n/a
name meaning : "lamb", "to bloom"
age : twenty - seven
gender / pronouns : cis woman / she/her
date of birth : september fourth
location of birth : istanbul , turkey
past residence : brooklyn , new york
past occupation : theoretical physicist
current affiliation : the rogues
moral alignment : lawful neutral
family : lucas campos ( fiancé ) , kamile aydin ( mother ) , alperen aydin ( father )
aesthetics :
playlist : on our own - bruno major , cruel world - active child
character inspiration : spencer reid - criminal minds , dana scully - the x files , lydia martin - teen wolf
𝚆𝙴'𝚁𝙴 𝙹𝚄𝚂𝚃 𝚂𝙲𝙰𝚁𝙴𝙳 𝚃𝙾 𝙰𝙳𝙼𝙸𝚃 𝚆𝙴'𝚁𝙴 𝙰𝙻𝙾𝙽𝙴
on september 4 1993 , no one was happier than kamile and alperen aydin . for their baby girl had entered the world . she was welcomed with a torrent of happy tears and zealous proclamations of never - ending love as well as veracious oaths to keep their baby daughter protected . they were never going to let her go .
and as they promised , talia grew up secluded from everything . she was never allowed beyond the boundaries of their house and their gates and fences were far too tall for little talia to even sneak a peek above . and her mother assured her , she wasn’t missing out on anything . but talia couldn’t help but wander , what was that noise ? an ice - cream truck that she’s read about ? or different types of birds and their different types of songs ? or other children , just like her ! who were willing to play tag and hide - and - seek ! she never got the answer
she was an only child and was home - schooled all her life , the burden of her parent’s expectations were particularly potent , especially about .. well , everythiing . they taught her four languages . besides her mother - tongue she was to also learn english , french , german and mandarin chinese . but the only sport dignified enough for the aydins was croquet . and then there was physics ; einstein’s theory of relativity , newton’s laws of motion , the big bang theory and so many more , biology ; natural selection and evolution , epigenetics , hormones and pregnancy and she was even taught about weedy and invasive plants , chemistry , maths and every other subject that was likely to cause the onset of depression in a pre - pubescent teenage girl .
but science , science she found a deep love for . it was interesting and magical even , it captivated her completely and her parents were happy to accomodate to her passion . giving her extra lessons and more posters and talia just couldn’t stop her fervid desire to know . and she rigorously dedicated herself to everything about it . to her it wasn’t about the grades or for the pride of her parents but for herself , for her own mind , to appease her own curiosity . it gave her freedom .
but as she grew older , the house looked smaller and the walls constantly felt like they were closing in . she was suffocating , she would feel claustrophobic in every room and she knew she had to get out of there before she lost her mind . so she snuck out , past their tall and frightening gates , past her cooking father and sleeping mother . as she took that first step , she felt like she had walked through a portal into another universe . there was such beauty , everywhere . and noise ! all kinds of noise , arguments and laughter , car honks and the roar of motorcycle engines . even the air smelt different . there was an aroma of food , new herbs and spices , it truly was an adventure she would never forget .
and on the way back , she had meticulously planned it you see , she was to leave at this time , grab these things , say this excuse and then leave at this exact time . otherwise , hell hath no fury like a mother scorned . but the cruel trickster fate , had different plans . on the way back she met a boy . a tourist , from brooklyn , new york . . he had honey eyes and dark skin and there was a strange new feeling that stirred in her stomach , like the flutter of butterflies or the whirring of a malfunctioning machine . they shook hands and exchanged names and talked for a while . but what stuck to her was where lucas was from , america , he answered , the land of the free . she left him with a smile on her face . despite knowing that her plan had been discarded and there was upcoming consequences to her actions , she couldn’t wipe the grin off her face . partly because of the boy and also because of the prospect of freedom . just a few oceans away , she felt like it was calling to her .
and , as she had predicted . her parents knew and they were enraged . and of course , talia had seen her parents angry before but this was new and alarming manifestation of their insecurities . they lectured her about the dangers of everything , of the world , of strangers , of cars , of air . ** trigger warning : parents being mean ? ** and they sent her to the basement , where it was cold and dark . and the only way that talia got through it all , with dry cheeks and a soaring heart , was because of that particular honey - eyed boy . the basement was not a new punishment . and the dark was an old friend . but what the darkness harboured was a stranger to her . her overactive imagination went haywire whenever she was in the basement -- fabricating monsters and gargoyles , winged - serpents and three - headed snakes . when she was down there , she could never sleep , she couldn’t stop shaking either . ** end trigger warning ** but she held on to the hope that she would meet that honey - eyed boy again and share the freedom he glorified in .
eighteen . she thought it was an age which warranted freedom and trust . but her parents only became more stern and harsh and utterly unyielding in the subject of talia’s freedom . she was to stay at the house , work with her father on his research project . and never leave . she cried about it a lot when she first heard the new guidelines of her existence . but her father explained it to her , her mother had lost one sister to disease , another in a traffic accident and a brother to alcohol . her father’s exact words were , “ she had failed in protecting her siblings but she wasn’t going to fail you . she loves you too much .” and talia understood , the only thing she could really do was accept it .
she worked with her father on his research project , he was a botanist and loved plants far more than the ordinary individual , he was always seeking for new plan species , even promising talia , that had he ever found one , he would name it after his daughter . for a while she was content , happy even but it didn’t last , it was never going to . she sought for something greater than the mundanities of a secluded lifestyle , she sought for love . and once she was curious about something , she wasn’t going to stop till her curiosity was satiated with answers .
at twenty - two , in the middle of the night , she worked up the courage , she stole her parent’s money ( not before leaving them a detailed letter mostly composing of apologies and proclamations of love and just a sentence or two asking them not to look for her ) , carrying nothing but a backpack and knowing absolutely nothing about the world , except from what she’s read , she headed to the airport . she caught a plane , brooklyn , new york . that was all lucas had told her . but that was all she needed . this was far greater than a boy now , this was about liberty .
at twenty - three , she had gotten a job , she had rented out an old and decrepit apartment and she was alone most of the time , but she couldn’t have been happier . she was free . what greater things were there than the liberty to choose , to be who you wanted to be . then love . love found her and wrapped her up . as though bound by the red string of fate , she and lucas found each other . and they grew to trust each other and love each other and three years on , they were engaged .
and as a little present for her , a pre - honeymoon , lucas had arranged for them to go on a roadtrip . first stop , delaware . she had heard about midway speedway park ( for go - karting and mini golf ! ) and jungle jim’s ( a seasonal water park with games ! ) from a co - worker and talia was completely enamoured with the idea of go - karting ! and water - slides ! she didn’t stop talking about it for days . and lucas , who knew his fiancé more than she knew herself , was enamoured with the idea of making her happy .
but as soon as they got there , something changed . like a flip had been switched off and they were hit with darkness . chaos and anarchy ensued and the worst flaws of human nature emerged . people were cruel and merciless and talia , who had been secluded for a great chunk of her life and who was taught to think meticulously and methodically , found it difficult to find the calculations in chaos . and it sent her brain into haywire . * trigger warning : anxiety attacks * she was anxious and panicked and most of the nights were spent with lucas trying to help her breathe . * end trigger warning *
as long as she was with lucas , she could do it , she could keep going . but a few months in this new reality , they were separated . lucas had protected her , begged her to run while he tried to fight for his life . and talia , who was ready to stay , ready to leave earth with the love of her life , ran . and she ran and ran , until she couldn’t breathe anymore , until her legs collapsed from under her , the screams of her soulmate still ringing in her ears . that was when she found the rogues , or rather the rouges found her . and everyday after that , all she could her was her mother’s warning voice , curiosity killed the cat .
but she wasn’t going to give up on lucas . there was a chance he could still be a live . she thought about everything methodically and meticulously , applying principles of science and laws of nature but love , love was an exception -- lucas was an exception . and that night she decided that nothing could stop her from finding him . yes , curiosity killed the cat . but satisfaction brought it back .
𝚆𝙷𝙰𝚃 𝙸𝙵 𝚃𝙷𝙴 𝚆𝙾𝚁𝙻𝙳 𝙸𝚂 𝙰 𝙷𝙾𝙿𝙴𝙻𝙴𝚂𝚂 𝙿𝙻𝙰𝙲𝙴
a kid , actually though . because she was secluded from everything , she is fascinated about everything . she is also very in love with earth , she wants to protect it and capture as much of the old world as possible . so she usually carries around a notebook and a pen , asking people their stories and what their life was . she literally stacked up on disposable cameras and a lot of film when she had walked past it . so she’s taking picture with those too sometimes .
she’s abrasive sometimes . she didn’t have a lot of interactions with other people . and sometimes she says things without thinking , and even though she had some practice , she might still say something insensitive because she didn’t know it was insensitive --- basically , she’s emotionally stupid . and she doesn’t know much about feelings . she feels them and she knows she feels them but its all still quite new to her .
she’s a compliant rogue , she does what she’s asked and abides whatever rules they’ve established . but she’s quite annoying , she calls people out on their stupidity and always kinda like thinks she’s better than everyone . she’s definitely not shy about highlighting people’s mistake and she loves LOOOVES telling people ‘ i told you so ’
she’s very physically weak . she doesn’t have any arm strength and she’s moderately good at running . but yeah , nah . she can’t fight for shit .
𝙰𝙽𝙳 𝚆𝙴'𝚁𝙴 𝙾𝙽 𝙾𝚄𝚁 𝙾𝚆𝙽
maybe the rogue who found her all sad and stuff ? and since they helped talia when she was such in a vulnerable state , she really trusts them and confides in them a lot ? and she always keeps them close because she’s scared of them getting hurt or them being separated .
also , i’m sending a wc form to the main but is there anyone who mayaps want to write lucas ?? maybe he survived and found the hartly compound ??? ( of couursee you can change their name and their personality , everything is basically up to you ! )
someone who’s helping her toughen up ????
someone who is annoying but she low - key loves and relies on ?? or vice versa !!
a little sibling / big sibling kind of dynamic !!
umm , idk but definitely lots more !!! i’m one - hundo percent up for brainstorming !!!
TLDR
been sheltered from everything most of her life , got separated with her fiancee , found the rogues , not giving up on finding her fiancee , she’s very annoying and says things without filtering them in her mind first , she’s kinda uptight , loves to show - off what she knows but loves to tell people ‘ i told you so ’ even more XD
#[[ this is a new benchmark guys .... this is not only 1000 but 2000 worDS adsfhjk ]]#[[ tldr is at the bottom !! hehhe ]]#miseriaintro
3 notes
·
View notes
Link
How old is your dog? Maybe it was born four years ago. At that age, a human would still be a kid. Your dog, however, acts like an adult. You might have heard that to get a dog’s “biological” age, just multiply its age in years by seven. That would make your dog equivalent to a 28-year-old human. But that would probably be wrong, a new study shows. Your dog’s actually more like a 53-year-old human.
Multiplying a dog’s age by seven doesn’t really work, says Matteo Pellegrini at the University of California, Los Angeles. He studies bioinformatics — a research field that uses computers to analyze biological data. He was not involved in the study. On the surface, he notes, multiplying by seven makes sense. On average, people live seven times longer than a dog. So, he notes, “It’s just dividing the human lifespan by the dog lifespan.”
But there are problems with that simple computation. After all, a one-year-old baby is just learning to walk. A one-year-old dog is adult enough to have puppies of its own. This is because species develop at very different rates. Aging doesn’t even happen at the same pace over an individual’s life. “When you’re a newborn, your body is changing very quickly,” Pellegrini explains. “It slows down over time.”
Explainer: What is epigenetics?
Fortunately, dogs and humans both hit very similar developmental milestones. We’re babies (or puppies), then kids (or, er, still puppies), then adolescents and then adults. As we go through those stages, our DNA also undergoes a change. The molecule that carries genetic instructions stays the same. But over time, this DNA acquires or loses tiny chemical “tags.” These are called epigenetic changes. The tags, known as methyl groups, act like little switches that can turn on or off particular genes in that DNA.
Explainer: What are genes?
The genome is the complete set of the genes present in an organism’s DNA. “Think of taking a … marker and start drawing in different places in the genome,” says Elaine Ostrander. She studies genetics at the National Human Genome Research Institute in Bethesda, Md. The marks determine how different genes in the DNA get used to make proteins. A complete set of the methyl marks on DNA is called a methylome (METH-el-ohm).
As an animal ages, so does its methylome. How it changes is very predictable. Most adolescent humans will have DNA marks that are in one pattern, while humans nearing 65 or 70 show a different pattern of DNA marks. The same is true for dogs. If the epigenetic markings of humans and dogs at different ages were compared against one another, would there be “places that were kept getting marked up in the same way?” Ostrander and her colleagues wondered. If there were, would finding that pattern in one species point to an equivalent age in the other? Comparing patterns that way might allow people to get at the “real” age of a dog.
To test this, her team looked at the methylomes from 104 purebred Labrador retrievers. Her team has been collecting DNA samples from dogs around the United States since 1993. “I picked Labs because they can live a long time [for dogs], and we were able to pull [the DNA] out of the freezer,” Ostrander says. “We were able to take DNA samples from Labs that were anywhere from a few months old to 16 or 17 years old.” The researchers compared the dog methylomes to methylomes of 320 people between the ages of 1 and 103.
And it worked. By comparing the patterns, they scientists found they could figure out how dog years relate to human years.
6 notes
·
View notes
Link
'In the study, participants were given a growth hormone and two diabetes medications. Scientists then monitored the test subjects’ epigenetic clocks, to understand the effect on how they aged.... 'As people age, chemical modifications or tags are added to people’s DNA, and those change throughout their lives, so by looking at those tags a person’s biological age can be measured.... 'Professor Horvath then looked at four different measures of the epigenetic clock to understand the differing ages of each of the patients. And he found that every one of them had reversed significantly – so significantly that he is optimistic about the results, despite the limited number of participants....'
1 note
·
View note
Text
Threads, worms and science communication
I thought I had written my last post about epigenetics. But then came along some ‘worms’ and I had to write another one.
I have written about worms once before on the Making Science Public blog, in the context of science communication. And this blog post too will reflect on worms in the context of science communication, but in a slightly different context.
In 2014, when I wrote my first worm post, twitter was eight years old but still evolving. Now we live in a twitterverse where science communicators can use ‘threads’, launched at the end of 2017, to knit together tweets into a science story. Some of the best science communication now happens in ‘threads’. And this is what happened on this occasion.
So what am I talking about? On 6 June a research group working in Tel Aviv around Oded Rechavi published an article in Cell entitled “Neuronal Small RNAs Control Behavior Transgenerationally”. The gist of the article was this, as summarised in the abstract: “In Caenorhabditis elegans nematodes, parental responses can transmit heritable small RNAs that regulate gene expression transgenerationally. In this study, we show that a neuronal process can impact the next generations.”
So the hero of the study is C. elegans, the worm of wonder I had talked about in my previous blog, but not in the context of epigenetics. What has this to do with epigenetics?
Worms and epigenetics
Epigenetics is a new field in genetics and genomics, which came to prominence around the year 2000 and about which I have written since 2013. It is still finding its feet and it is still struggling with definitions and concepts. One of the most contested notions is that of ‘transgenerational epigenetic inheritance’. This phenomenon has been observed in plants, worms and some rodents, but there is, as yet, no clear evidence that it exists in mammals/humans.
In an important article summarising some myths surrounding epigenetics, Edith Heard, a renowned epigenetics researcher, pointed out: “Since the human genome was sequenced, the term ‘epigenetics’ is increasingly being associated with the hope that we are more than just the sum of our genes. Might what we eat, the air we breathe, or even the emotions we feel influence not only our genes but those of descendants?” This is at the core of speculations surrounding epigenetics and transgenerational epigenetic inheritance.
The worm study that caught my eye has to be seen in this context. It was announced under headlines such as “Worm parents pass on behaviours epigenetically to offspring”.
Even one of the most sceptical observers of research into transgenerational epigenetic inheritance, Kevin Mitchell, tweeted: “Some real transgenerational epigenetics… (in worms)”.
He also retweeted a ‘thread’ by the group leader, Oded Rechavi, which was a masterpiece of science communication, both verbal and visual – indeed, threading them both creatively together. We’ll come to that in a minute.
Epigenetic worms in the media
How was the study reported in the ‘mainstream media’? I had a quick look and found only four articles, all published in Israeli news outlets. I tracked down a few more on Google News. Here are some of the headlines in the print media: “Israeli study: Nervous system can transmit messages to future generations”; “Israel study says neurons, not just DNA, can affect progeny’s fate”; “Researchers say nervous system passes info to kids”; “Worms help Israeli scientists rewrite basics of genetics”.
Two things struck me: first the use of anthropomorphic terms like ‘kids’, a type of language that can create the wrong expectations, and the claim that this study overthrows basic genetics. This claim came also up in an online article on Breaking Israel News entitled “Small worms help Israeli researchers disprove basic dogma of modern biology”. This article was, I believe, the press release for the Cell study and contains a great artistic illustration of worms. It was picked up by eurekalert, where Rechavi is quoted as saying “It’s important to stress that we don’t know yet whether any of this translates to humans”.
In the past, some epigeneticists and philosophers of science have made claims about epigenetics overthrowing basic biology, claims that were greeted mainly with scepticism by other scientists.
Three other online articles reported on the study, one published in The Scientist and dryly entitled: “Worm parents pass on behaviors epigenetically to offspring”; one in Psychology Today seeing it as ‘disruptive’ to biology and neuroscience, and one in the Big Think linking it to ‘reincarnation’ (tongue in cheek)! There may be more. But let’s get to the thread.
Twitter threads
According to Twitter, “[a] thread on Twitter is a series of connected Tweets from one person. With a thread you can provide additional context, an update, or an extended point by connecting multiple Tweets together.”
Here is a random example of a ‘thread’ – there are longer and more extravagant ones out there. This one is about Mayan hieroglyphics, which I loved (it’s linked to a fantastic podcast). You can click on it and see how it goes…
Knitting a science story using words and images
So, what about this worm thread then, which got 423 retweets and 1,156 likes? Rechavi posted it in the afternoon of 6 June, just after his lab’s article came out in Cell. It starts with a bit of a firework of emojis, hashtags, and hyperlinks.
The hashtag uses a well-established epigenetic metaphor. The accompanying image (called ‘a summary model’ in the thread) provides an overview of the ‘flow’ of signals/memory, I suppose, between neurons, germline and behaviour.
The second tweet congratulates all the team members and especially the artist who created the illustration for the press release Beata Edyta Mierzwa and reproduces the sci-art image. See here. I think the image represents a tree of life/arteries of life populated by worms with little epigenetic ‘tags’ or post-it notes attached to them… but I might be wrong.
The third tweet writes about long-standing speculations about inheritance and is illustrated by a photo of a Greek statue representing a thinker. It ends by stressing: “But transgenerational epigenetic inheritance is still extremely controversial, especially in mammals”.
The fourth tweet goes into this controversy in a bit more detail, saying: “The reason it’s controversial is that there’s no good mechanism that can explain it. It was hypothesized a long time ago (19th century) that the germline is isolated from the soma, and that somatic responses (including neuronal responses) cannot become heritable”. The work presented in the thread puts forward a candidate mechanism.
This tweet is illustrated with a photo of the front cover of August Weismann’s seminal book The Germplasm – a theory of heredity (published in 1893). This tweet relates to what some news articles reported, such as one in the Big Think that stated: “Rechavi believes this research pushes back against biological dogma (the ‘Weissmann Barrier’) claiming that heritable information is segregated from somatic influences.” This seems to be much less cautious than the tweet, probably because it’s based on the ‘press release’.
As there are 17 tweets in this thread, I won’t summarise each one of them. Let’s just say each tweet is a combination of text and image. It uses images of many types and genres, such a diagrams, various plots and scatter grams, cartoons, microscopy images, photos, and so on..
However, I want to point out one more tweet, not for its image but for its text, as it engages in anthropomorphic language (also used in some headlines) which might confuse people and make them think that transgenerational epigenetic inheritance has also been demonstrated in humans:
“MORE interestingly, if rde-4 great-grandchildren are derived from heterozygous great-grandparents that expressed rde-4 only in neurons, their chemotaxis behavior is much improved. The ancestors’ neurons control the behavior of the great-grandchildren”
Reception
The response to the thread was overwhelmingly positive with lots of tweets expressing congratulations. One tweet posted a compiled thread on something called ‘threader’, which you can read here. And there is an ‘unroll’ here on a thread reader.
You can also read the compiled text of the tweets on Reddit. There are only a few comments related to that text, but three of them are interesting. One continues the anthropomorphising of the worms and creates a great metaphor: “Grandpa worm sent a zip file of worm behavior instructions to grandbaby worm”.
Two others jump into a controversy surrounding transgenerational epigenetic inheritance that been reverberating through popular science for a while: “Lamarckism wasn’t as wrong as we thought.” While somebody else says (in a longer comment): “I definitely don’t want to dismiss this but I dont know if I’d call this ‘heritable’ in the same way DNA is heritable.” Discuss!
A new type of science communication
The worm study has provided me with a great opportunity to learn about threads and to alert readers of the Making Science Public blog to the fact that there are now new ways of making science public, using the old art of intertwining words and images (see also GIFs), texts and illustrations, art and science.
There is no yet a lot of research out there examining the use of twitter threads for science communication. I found one SciCommthreads hashtag on threads, but that’s about it. There must be more!
On a different but related note: I am looking forward to a new little project with Aleksandra Stelmach and Alan-Miguel Valdez examining the power of images in the construction of popular notions of transgenerational epigenetic inheritance.
Image: Wikimedia Commons: C. elegans, stained
The post Threads, worms and science communication appeared first on Making Science Public.
via Making Science Public http://bit.ly/2KOJ5Q7
2 notes
·
View notes
Text
Identical twins carry genetic modifications no one else has | Science


Identical twins are living proof of how genetics shapes our looks and traits. Now, researchers have found they carry a molecular signature on their DNA that no one else has—one that becomes fixed in their cells early in development and stays with them into adulthood.
This signature doesn’t seem to influence a twin’s health, but it could offer insights into how identical twinning happens. “It is a starting point” for solving “what is really an enigma,” says Jenny van Dongen, a twin genetics researcher at Free University (VU), Amsterdam. The signature could also be used to test whether a person had a “vanishing twin,” an identical twin that died in the womb.
Identical twins occur when an egg splits soon after it is fertilized, resulting in two embryos with the same DNA. Scientists have some idea how fraternal twins, which result when two eggs are fertilized, happen: Both genetics and age may tip ovulating women toward producing two or more eggs at once. But experts don’t really understand what factors lead to identical twins, which make up about four of 1000 births.
An international team led by van Dongen and VU twin genetics researcher Dorret Boomsma looked for clues in what’s known as the epigenome. Patterns of chemical tags called methyl groups glom onto genes, turning them on or off. (Such epigenetic changes are responsible for everything from enabling Peruvians to live at high altitudes to helping the placenta develop.)
Using blood and cheek cell samples, the researchers scanned the epigenomes of more than 3000 identical twins, as well as a comparable number of fraternal twins and some twins’ parents. They looked at 400,000 different places on each person’s genome. About 800 locations had differences in methylation that set identical twins apart from everyone else, the team reports today in Nature Communications. “It’s likely something established very early on that is propagated to subsequent cells,” van Dongen says.
Some of the methylated or unmethylated spots made sense, such as tags on genes involved in cell adhesion that might influence how easily a fertilized egg splits into two embryos. But changes in other locations, such as the ends of chromosomes, don’t have an obvious explanation. These regions have been associated with aging, yet identical twins’ life spans are similar to other people’s.
Understanding the epigenetic changes and whether they are a cause or effect of an egg splitting will require lab studies using embryolike structures made from stem cells or early animal and human embryos (with appropriate ethical review), says Jeffrey Craig, a twin researcher at Deakin University, Geelong Waurn Ponds. For now, the findings offer “fascinating insights into potential mechanisms in [identical] twinning,” says twin researcher Nancy Pedersen of the Karolinska Institute, who like Craig wasn’t involved with the research.
The work could shed light on some rare disorders involving epigenetic changes, the VU-led team says. The growth disorder Beckwith-Wiedemann syndrome, for example, is more common in identical twins than in single births. It could even help firm up a hypothesis that such disorders only occur in identical twins, which would be “big news if proved true,” Craig says.
An epigenetic test might also be useful to determine whether a person once had an identical twin that vanished in the uterus, perhaps because it didn’t have enough room or nutrients to grow. Sometimes a twin fetus appears in an ultrasound before vanishing, but other times it may be absorbed without leaving a trace. As many as 12% of pregnancies start out as multiples (including fraternal twins), according to some estimates, but only 2% of twin pairs survive.
Using a separate data set, the epigenetic signature could predict whether someone was an identical twin in 70% to 80% of cases, van Dongen says. With data from a large enough group of people, the test would get even better, she says, and it could also help “predict the exact rate” of vanishing twins. That figure would be useful not only for researchers, but also “of broad interest” to twins themselves and to families who are mourning the loss of an identical twin, Boomsma says.
New post published on: https://livescience.tech/2021/09/29/identical-twins-carry-genetic-modifications-no-one-else-has-science/
0 notes
Text
Biomed Grid | Hybrid Rice Technology: An Overview
Introduction
The prevalence of today’s environmental contaminants is unprecedented. The Toxic Substance Control Act Chemical Substance Inventory lists over 85,000 chemicals in the U.S. market [1]. Adapting to persistent organic pollutants (POPs) exposures is difficult, particularly if adaptive responses require novel genetic variation [2]. An individual’s fitness is influenced by the sum of all stressors, which can be additive, antagonistic, and/or synergistic [3-5]. Devising effective long-term risk-assessment and mitigation strategies is complicated further by human-induced selection pressures on natural populations (climate change, habitat conversion…), jeopardizing organisms’ abilities to survive and adapt to acute and chronic environmental contaminants. Identifying and quantifying such adaptations in individuals and populations in their natural setting is difficult. Yet, technological advances in next generation sequencing (NGS) and transcriptomics provide an opportunity to infer statistically and biologically relevant information within complex but manageable data sets.
POPs are toxic organic compounds resistant to environmental degradation. Most are anthropogenic, lipid-soluble and can cause adverse impacts on the environment and human health. Many POPs are industrial chemicals - solvents, pesticides, insecticides and fungicides, pharmaceuticals, polychlorinated biphenyls (PCBs), dioxins (PCDD), polyfluorinated dibenzofurans (PCDFs), etc. [6,7]. Due to their general use, POPs have high environmental concentrations; many are hydrophobic and bioaccumulate through the food chain due to their slow rate of metabolism [8-10]. Human and animal exposures are mostly through diet (90%), environmental exposures, and during embryo/fetal development via maternal deposition. Chronic POPs exposures can result in acute and chronic toxicity, demonstrated by developmental defects, chronic illnesses, and increased mortality [6]. Some are categorized as carcinogens [11] while many are endocrine disruptors [12-14] and may affect nervous, immune, digestive, urinary and reproductive systems [6]. Low-level exposure to POPs during critical fetal development stages can have a lifetime effect [12-15]. POPs concentrations in human serum increase with age and seem to be higher in females than males [16]. Evolutionary responses and adaptations to POPs have been documented since the mid-20th century; initially as pesticide resistance cases among invertebrates and plants [17,18], then among other natural populations, including vertebrates and mammals [19,20]. Although industrial toxic chemicals were provoking rapid evolutionary response among natural populations, their effects were mostly ignored.
To evaluate temporal and spatial exposure risks, government agencies study the most environmentally prevalent POPs by examining bioavailability and dose-response relationships in the laboratory setting [21]. Such assessments are limited within relatively static laboratory settings. For organisms exposed to a mixture of POPs, the adverse effects are assumed to be additive as mixtures of POPs can result in synergistic effects, enhancing the toxicity of each compound [3-5]. However, responses to POPs are complex and should be carefully considered when quantifying evolution’s role in mechanisms of sensitivity and resistance to complex mixtures of pollutants within and between natural, genetically variable populations [22,23]. Evolutionary changes due to a chronic POP exposure can be quantified in just a few generations [24,25] and include physiological, genetic and epigenetic transgenerational effects influenced by natural selection: studies utilizing natural populations report phenotypic and molecular consequences, including increased mutation rates, receptor desensitization, phenological traits and epigenetic effects [26,27]. While some exposure effects are adaptive, others reduce population size, causing inbreeding depression or genetic drift [28,29] and decreased genetic variation, compromising the ability to adapt to future stressors. Adaptation to POPs can evolve at a cost, such as increased subsequent sensitivity to oxidative stress [30]. Risk assessment of environmental exposures to complex chemical mixtures should not be limited to often oversimplified and outdated controlled laboratory bioassays focused on robust acute and chronic toxicity endpoints such as survival and reproductive viability of model organisms lacking inherent genetic variation.
The post-industrial age of overwhelmingly complex chemical exposures coupled by recent technological advancements in NGS methodology allows for the utility of diverse non-model organisms within an evolutionary context in public health risk assessment. Scientists started using population genomic approaches to study natural populations lacking robust genomic resources a decade ago. Given that genome sequences of non-model organisms are accumulating at an unprecedented pace, with 234 animal and 319 contig genome assemblies currently in development [31], technological advancements in high-throughput sequencing from non-model organisms generate critically important data to infer complex genotype- environment interactions in a natural setting. Population genomic studies target the underlying variation found in the DNA among individuals and populations. They also utilize genome-wide sampling of sequence variation under the premise that demography and the evolutionary history of populations affect neutral loci similarly, whereas loci under selection will be affected differently [32]. Data of multiple, although individually analyzed, loci from natural populations enables studies of the genomic evolution, acclimatization, and adaptation to strong selective pressure such as pollution exposure in natural environment. Such data sets can yield insights about population processes and variance of allelic diversity within and between populations. The advantage is that an a priori choice of biomarkers of an adverse effect is not necessary, which is important when organisms are exposed to complex chemical mixtures in their natural environment and biomarkers of exposure are not well established [33]. Moreover, adaptation-related projects looking for genomic signatures of selection within and between natural populations can take advantage of established adaptative phenotypes to environmental gradients.
A typical population genomic study platform consists of sequencing strategy design, generation of sequence data, mapping of sequence reads to the assembly, genotyping, and population genetic/ molecular evolutionary analyses. A sequencing strategy includes depth of coverage and utility of individuals vs. pooled samples, number of individuals per population and number of populations, gender, and the need for outgroup species. High-throughput genotyping of millions of markers simultaneously is available for model organisms. Whole transcriptome sequencing has advantages but is expensive since multiple individuals need to be sequenced to represent the population adequately. Moreover, the analysis of short sequence reads depends on an incomplete reference genome to which sequence reads are aligned. Such sequencing platforms generate massive data sets, meaning that inferring statistical and biological relevance can be challenging. Although most non-model natural population genomes are not yet sequenced, reasonably robust, lower throughput, custom platforms are available. Expressed sequence tags (ESTs) have been developed and used as genome-wide gene expression data within and among natural fish populations historically exposed to chemical pollution. New sequencing technologies such as Roche (454) FLX, Illumina Genome Analyzer, ABI SOLiD, and HeliScope sequencing [33] can be used to sequence thousands of transcripts quickly and cost-effectively, making transcriptomic studies possible in virtually any species. Transcriptomics and population genomics can influence studies of diverse species and populations that ask important ecological, physiological, and evolutionary questions with respect to pollutant exposure. High-throughput sequencing allows enough sequencing depth to gain an adequate representation of all the expressed transcripts and has been used for whole transcriptome sequencing [34]. The coding sequences of the expressed transcripts can be analyzed for mutations, altered splice sites, and protein polymorphisms.
Whole transcriptome approaches can identify significant changes in gene expression that are biologically important, but the genomic basis underlying altered patterns of gene expression is mostly unknown. Once many genetic markers became available, population geneticists began scanning genomes for reduced nucleotide diversity, extended linkage disequilibrium, or regions of homozygosity as potential selection signatures. This approach requires that large numbers of loci and genetic markers are statistically analyzed for non-random patterns [35]. At loci under selection, local adaptation and directional selection should reduce genetic variability within populations and increase variation among populations. Loci used in population genomics studies include microsatellites, single nucleotide polymorphisms (SNPs), amplified fragment length polymorphisms (AFLPs), randomly amplified polymorphic DNA (RAPDs), and sequences (e.g., whole-genome sequences). Many genetic markers can be generated without a sequenced genome relatively easily and genomes can be mined without measuring phenotypes, allowing for sampling of individuals without knowing their breeding history. For example, SNP analysis can be performed by mining EST data collections for putative SNPs [36]. If the ESTs were derived from multiple individuals (and multiple populations of interest), putative SNPs can be identified in sequence alignments. Conveniently, any SNPs identified as important through population genomic approaches are already associated with a sequenced gene transcript.
Custom SNP genotyping platforms provide efficient genotyping of thousands of individuals, resulting in biologically important sequence variations from multiple populations. The advantage of AFLPs and RAPDs is that, again, no prior sequence information is necessary. The drawback is that AFLP and RAPD analyses depend on high-quality DNA and provide only dominant markers, so heterozygotes cannot be directly measured, and generated markers are often anonymous or in a nonfunctional area of the genome. Genome-wide association studies (GWAS) are challenging with non-model organisms because they lack an abundance of readily available, high quality, sequenced genomes, and genome-wide genotyping data. To improve the use of NGS data obtained from non-model organisms, new imputation methods such as Link Imputer [37] help infer missing genotypes and facilitate the analysis of non-model organisms sequencing data.
Susceptibility to toxic substances depends on the organism’s genotype and interactions with the polluted environment. Natural populations inhabit spatially and temporally uncontrolled environments where pollutants are present as complex mixtures, so the challenges of “omics” technologies with natural populations are the challenges of basic biological research: which data is biologically meaningful? Considering cost, challenging sample material, lack of biological replicates, and complexity of genotype-environment interactions, choosing an optimal method is difficult. Yet, instead of a single gene, protein, or polymorphism, one can analyze thousands of genes, proteins, and polymorphisms utilizing biologically realistic, genetically variable individuals, extrapolating a population response rather than a strain-specific one. Complementary “omics” approaches offer a better understanding of biological effects at multiple levels: model-organisms are used to study many important questions in biology, population genomics provide insights into the sequence variations that govern differences in gene expression and protein polymorphisms, while transcriptomics provides insights into altered protein levels. A focused approach, using known POP inducers and inhibitors present in such mixtures and analyzing responses of cellular and tissue targets, can provide insights into mechanisms of toxicity seen in natural populations and help devise a more comprehensive risk assessment strategy. Thus, just as the integration of laboratory and field studies improves our knowledge of genes and proteins, so is the integration of laboratory and field studies critical for “omic” approaches and public health risk assessment modeling.

Read More About this Article: https://biomedgrid.com/fulltext/volume5/hybrid-rice-technology-an-overview.000865.php
For more about: Journals on Biomedical Science :Biomed Grid | Current Issue
#biomedgrid#american journal of biomedical science & research#Biomedical Science and Research Journals#Health science Journal of Open access
0 notes
Text
Aftershocks Notes
Nadia Owusu.
“I asked my father what an aftershock was. He said there are tremors in the earth that follow an earthquake. They are the earth’s delayed reaction to stress.”
“...an English girl with green eyes, wants to know why my mother and I are different colors. There is no malice, only curiosity, in her voice, but I feel embarrassed. I can only say I don’t know why. As I return to my seat, my face burns”
“At lunchtime, Miss Rossi, my teacher, sits next to me and asks if I enjoyed spending time with my mother...How do I tell her about the trembling that leads to rippling, then to violent rupture; to whole lives and whole cities disintegrating; to piles and piles of rubble, to displacement and exile? How do i tell her that a day that begins with pancakes for breakfast can end in disaster, that, in an instant, an earthquake or a mother can arrive ad change everything? How do I tell her that even when the earth stops shaking, cracks in the surface spread silently? Pent-up forces of danger and chaos can be unleashed at any time. I don’t know how to explain any of this, so I tell her I am afraid of the after-shocks”
The next chapter is her explaining the answers as she fills out a “ refugee resettlement form” it isn’t as simple as one language, one hometown, etc.
On her fathers death - “Grieving, I learned, was a process of story construction. I needed to construct a story so I could reconstruct my world. There were decisions to make about what to put in and what to leave out.”
“I believed the vibrations and alarm were caused by an instrument in my brain. My seismometer, I called it. I called it that because I had, since my mother arrived with an earthquake when I was seven, been obsessed with earthquakes and the way we measure them, the ways we try to understand the size and scale of impending disaster. When I say I believed in my seismometer I mean it was an irrefutable conviction. I came to know my seismometer was there in the same way I knew I had a mole below the right corner of my bottom lip...”
“Authors - Chinua Achebe, Wole Soyinka, Nurrudin Farah, Mariama Ba, Kofi Awoonor. Music - Fela Kuti, Miriam Makeba, Salif Keita, Youssou N’Dour, E.T. Mensah and the Tempos Band”
“A study from New York’s Mt. Sinai Hospital found that genetic changes stemming from the trauma suffered by Holocaust survivors were capable of being passed on to their children. Our genes change all the time when chemical tags attach themselves to the DNA and turn genes on or off. The study found that some of these tags - found in the genes of those survivors - were also found in their children. The changes led to an increased incidence of stress disorders. This passing down of environmentally altered genes is called epigenetic inheritance”
“Some things are out of the god’s control. And some matters require more from us than hope or prayer. They require us to see and support one another. They require us to defend on another. We must all, in the end, make peace with that.”
“Code switching is dancing between vocal styles and rhythms...This dance is part celebration - of the richness, intricacies, and blurry borders of our cultures...And this dance is also part survival mechanism. ...there is pressure to sound whiter”
“Sometimes I think my memories are more about what didn’t happen than what did, who wasn't there than who was. My memories are about leaving and being left. They are about absence”
“I feel something like a relief, but heavier. It is like taking off a winter coat in a hot room only to find that I cannot get out of the wool sweater underneath”
Also she was going to university a few blocks away from the world trade center, and was on a train heading there the morning of the attack...crazy.
“Doing things I’m very bad at makes me sad about all the things in the world I will probably never really understand, like electricity and Einstein’s general theory of relativity”
0 notes
Quote
A study from New York's Mount Sinai Hospital found that genetic changes stemming from the trauma suffered by Holocaust survivors were capable of being passed on to their children. Our genes change all the time when chemical tags attach themselves to the DNA and turn genes on or off. The study found that some of these tags -- found in the genes of those survivors -- were also found in their children. The changes led to an increased incidence of stress disorders. This passing down of environmentally altered genes is called epigenetic inheritance.
Nadia Owusu from Aftershocks
0 notes
Link
When it comes to career paths, worker ants split into castes: Some tackle defense, others forage for the colony. But these roles aren’t predestined. An ant’s career trajectory is influenced by factors in its environment early on in life.
Now, a study reveals one way those environmental factors play out. A protein called CoREST acts like a molecular switch in Florida carpenter ants (Camponotus floridanus), researchers report November 12 in Molecular Cell. By toggling it, big workers fated to be soldiers can be reprogrammed to do the job of their smaller, forager sisters.
Brawny warriors, called majors, and foraging, nursery-tending workers, called minors, share nearly identical sets of DNA. So researchers have looked for epigenetic influences, chemical tags on DNA and associated proteins that can manipulate how genes are read, to explain the different behaviors.
“And that’s what we found,” says Shelley Berger, a molecular biologist at the University of Pennsylvania Perelman School of Medicine. “It’s the first epigenetic mechanism that’s been found in ants to regulate behavior in the brain.”
The new study highlights that even highly specialized social insects retain substantial flexibility and responsiveness to their environment, says Beryl Jones, an evolutionary biologist at Princeton University not involved in the research. “This is likely another important facet of the great success of social insects,” she says.
Berger’s team had previously shown that injection of a chemical, trichostatin A, that helps unwind tightly packaged DNA could reprogram the majors to behave like minors (SN: 12/31/15). But it wasn’t clear what genes trichostatin A was influencing, or how far along in their development the ants could still switch jobs.
In the new study, Berger and her colleagues injected trichostatin A into the brains of worker ants either zero, five or 10 days after the ants emerged as adults from their pupal stage. These injections could reprogram major workers up to five days into their adult stage, but if administered later, the ants’ roles were already cast and it was too late to change them.
youtube
Warrior ants, called majors, that are reprogrammed with an injection of a chemical called trichostatin A (left) forage for food more frequently than natural major workers. Like their foraging and nursery-tending worker kin, called minors, (right), the warrior ants regularly forage at a sugar water pool at the center of an arena.
When the team analyzed gene activity during this five-day window of sensitivity the reprogrammed major workers were producing more CoREST protein than those not injected with trichostatin A. CoREST represses enzymes that break down juvenile hormone, a hormone normally elevated in minor workers, but not in majors, the team found. Boosting CoREST made the warriors more like their minor foraging brethren — with less enzyme production and more juvenile hormone.
CoREST has long been linked to neurological development, but this is the first time it’s been shown to influence behavior. The protein is found in a wide range of animals, including mammals. Given its ubiquity, CoREST may even play a key role in human biology, Berger says.
“Certainly there are going to be differences between humans and ants,” she says. “But what if CoREST is a really important epigenetic regulator of behavior in humans during early life and even early adolescence?”
Berger next wants to investigate epigenetic changes in ants’ individual brain cells, to see if specific cell types are key to the link between CoREST levels and ant behavior. Studies like these that unveil the genetic underpinnings of insect societies make for an exciting time to be studying social insects, says Daniel Kronauer, a biologist at the Rockefeller University in New York City not involved with this research. “People have wondered about the mechanisms underlying [ant] caste development and division of labor for so long, and it feels like we’re finally on the verge of understanding them,” he says.
17 notes
·
View notes
Text
92 Truths
I was tagged by @saisakurano5 , @noa-noa-noa, and @youngksnose! )Thank you very much I’m sorry for being so slow, I really wanted to do this!)
rules: once you’ve been tagged, you are supposed to write a note with 92 truths about you. at the end choose 25 people to be tagged!
last:
drink: water
phone call: my mum
text message: My group partner- “Lets edit it by tomorrow”
song you listened to: “The Greatest Show”- after all these years, I still have a crush on Zac Efron (no but it’s a really good movie)
time you cried: A couple days ago (this semester is overwhelming ugh)
have you ever:
dated someone twice: No
been cheated on: No
kissed someone and regretted it: No
lost someone special: No
been depressed: .....yeh
been drunk and thrown up: I wasn’t drunk but i did drink and it was terrible and i had food poisoning at the same time NEVER AGAIN
in the past year have you:
made a new friend: Yeah! online!
fallen out of love: No
laughed until you cried: Yeah
met someone who changed you: i don’t think so
found out who your true friends are: not particularly
found out someone was talking about you: no
general:
how many people on tumblr do you know in real life?: like met irl? a few!
do you have any pets?: i had a goldfish when i was like 7
do you want to change your name?: not really
what time did you wake up this morning: 5:30 TT I left early to study for my 8am class, and I couldn’t sleep
what were you doing last night: Reading a very boring paper about epigenetics (fascinating! (a joke))
name something you cannot wait for: BLACK PANTHER!!!!!
have you ever talked to a person named tom?: Yeah I had a coworker named that! He was super nice!
what’s getting on your nerves right now: Classes
blood type: I don’t know! I really want to but I’m anemic so I don’t think I can donate blood :(
nickname: its embarrassing!
relationship status: single
zodiac sign: Pisces
pronouns: she/her
favorite show: I don’t really watch alot nowadays! My favorite Kdramas are Goblin, W and Healer, and rn I’m watching ANTM and Star but they are more like guilty pleasures haha
college: In my last semester and crying everyday lmao!
hair color: black
do you have a crush on someone: No
what do you like about yourself: BE POSITIVE! ugh I can have a good work ethic!
firsts:
first surgery: I got my tonsils out when I was like 2
first piercing: I don’t have any piercings actually, not even my ears
first sport you joined: HA SPORTS thats a good one (nah i’m very clumsy and don’t really like sports)
first vacation: Jamaica when I was really little
first pair of sneakers: Probably something from payless? Idk haha
right now:
eating: pizza
drinking: water
i’m about to: play mario kart!
listening to: The new Japanese release! ITS SO GOOD UGH
want kids: At some point in the future, sure!
get married: um sure!
career: Having one with benefits!
which is better:
lips or eyes: eyes, nose, lips
hugs or kisses: hugs!
shorter or taller: I think I would like taller but it doesn't matter
older or younger: older, but it doesn't really matter
romantic or spontaneous: romantic!
sensitive or loud: sensitive?
hookup or relationship: relationship haha
troublemaker or hesitant: hesitant?
have you ever:
kissed a stranger:No
drank hard liquor: I had a shot once- I don’t think alcohol is for me like if you have to have a bunch for it to not taste awful that’s not so great
lost contacts/glasses: NO! Them things expensive!
sex on first date: No
broken someone’s heart: I hope not
been arrested: no
turned someone down: no
fallen for a friend: no
do you believe:
in yourself: I’m trying to I really am TT
miracles: YES!
love at first sight: maybe like attraction, but you need to get to know somebody before you fall in love I think.
25 is too many! Tagging @iloveniallthenandnow @my-little-talks @day-zzed @ultamber @upside-downlemur You don’t have to do it of course and anyone else who wants to go ahead! this was fun!
4 notes
·
View notes
Text
To figure out your dog’s ‘real’ age, you’ll need a calculator
How old is your dog? Maybe it was born four years ago. At that age, a human would still be a kid. Your dog, however, acts like an adult. You might have heard that to get a dog’s “biological” age, just multiply its age in years by seven. That would make your dog equivalent to a 28-year-old human. But that would probably be wrong, a new study shows. Your dog’s actually more like a 53-year-old human.
Multiplying a dog’s age by seven doesn’t really work, says Matteo Pellegrini at the University of California, Los Angeles. He studies bioinformatics — a research field that uses computers to analyze biological data. He was not involved in the study. On the surface, he notes, multiplying by seven makes sense. On average, people live seven times longer than a dog. So, he notes, “It’s just dividing the human lifespan by the dog lifespan.”
But there are problems with that simple computation. After all, a one-year-old baby is just learning to walk. A one-year-old dog is adult enough to have puppies of its own. This is because species develop at very different rates. Aging doesn’t even happen at the same pace over an individual’s life. “When you’re a newborn, your body is changing very quickly,” Pellegrini explains. “It slows down over time.”
Explainer: What is epigenetics?
Fortunately, dogs and humans both hit very similar developmental milestones. We’re babies (or puppies), then kids (or, er, still puppies), then adolescents and then adults. As we go through those stages, our DNA also undergoes a change. The molecule that carries genetic instructions stays the same. But over time, this DNA acquires or loses tiny chemical “tags.” These are called epigenetic changes. The tags, known as methyl groups, act like little switches that can turn on or off particular genes in that DNA.
Explainer: What are genes?
The genome is the complete set of the genes present in an organism’s DNA. “Think of taking a … marker and start drawing in different places in the genome,” says Elaine Ostrander. She studies genetics at the National Human Genome Research Institute in Bethesda, Md. The marks determine how different genes in the DNA get used to make proteins. A complete set of the methyl marks on DNA is called a methylome (METH-el-ohm).
As an animal ages, so does its methylome. How it changes is very predictable. Most adolescent humans will have DNA marks that are in one pattern, while humans nearing 65 or 70 show a different pattern of DNA marks. The same is true for dogs. If the epigenetic markings of humans and dogs at different ages were compared against one another, would there be “places that were kept getting marked up in the same way?” Ostrander and her colleagues wondered. If there were, would finding that pattern in one species point to an equivalent age in the other? Comparing patterns that way might allow people to get at the “real” age of a dog.
To test this, her team looked at the methylomes from 104 purebred Labrador retrievers. Her team has been collecting DNA samples from dogs around the United States since 1993. “I picked Labs because they can live a long time [for dogs], and we were able to pull [the DNA] out of the freezer,” Ostrander says. “We were able to take DNA samples from Labs that were anywhere from a few months old to 16 or 17 years old.” The researchers compared the dog methylomes to methylomes of 320 people between the ages of 1 and 103.
And it worked. By comparing the patterns, they scientists found they could figure out how dog years relate to human years.
Age isn’t just a number
How an age in years corresponds to a species’ biological age changed over time, they found. “You don’t see a … straight line,” Ostrander says. “What we see is this curve that levels out as humans get old and dogs get old.”
Explainer: What are logarithms and exponents?
Early in life, puppies develop much faster than people do. But as dogs get older, their aging curve begins to flatten. The scientists took this curve and developed a new mathematical equation to calculate a dog’s age. Sadly, it’s not as easy as multiplying by seven (that would just give you a straight line, not a curve). Ostrander and her colleagues published their new formula online July 2 in Cell Systems.
The new formula relies on the mathematical concept known as logarithms. And not just any logarithms, but those known as “natural” ones. To figure out a dog’s biological age, they say, multiply the natural logarithm of the dog’s age in years by 16. Then add 31.
Dog days
This graph compares Labrador retrievers at different ages with humans at different ages. It’s based on chemical tags on DNA and how they change over time in both species. It shows a 1-year-old dog is biologically closer to a 31 year-old human (second dot from left). A 4-year-old dog is like a 53-year-old human (third dot).

T. Wang et al/Cell Systems 2020
Such an operation is probably not math you can do in your head. (That’s okay, there’s an “ln” function on most calculators.) With this new formula, an 8-week-old puppy is roughly the same developmental age as a 9-month-old baby. A year-old dog corresponds to a 31-year-old adult human. And the 4-year-old dog from the beginning of the story is about comparable to a 53-year-old human. The new formula also lines up the average life span of a Lab (12 years) with the average 70-year human life span.
“I think it’s very cool they looked at the DNA methylation [patterns] and they compared them to match up the relative ages,” says Pellegrini. Other researchers will of course have to confirm the new findings, he adds.
And then there’s the issue of breed. The new study only used Labs. “There’s a big difference in longevity across dog breeds,” he notes. Smaller dogs, such as chihuahuas, can live close to 20 years. St. Bernards and other very large dogs might only live 10. Scientists will have to look at the methylomes of different dog breeds to see if they differ. They’re just going to have to study many more very good dogs.
To figure out your dog’s ‘real’ age, you’ll need a calculator published first on https://triviaqaweb.tumblr.com/
0 notes