#molecular structure
Explore tagged Tumblr posts
Text
Exploring the Marvels of Biological Macromolecules: The Molecular Machinery of Life (Part 3)
Proteins and Enzymes: Catalysts of Molecular Reactions
Proteins are the central players in macromolecular interactions. Enzymes, a specialized class of proteins, catalyze biochemical reactions with remarkable specificity. They bind to substrates, facilitate reactions, and release products, ensuring that cellular processes occur with precision.
Protein-Protein Interactions: Orchestrating Cellular Functions
Proteins often interact with other proteins to form dynamic complexes. These interactions are pivotal in processes such as signal transduction, where cascades of protein-protein interactions transmit signals within cells, regulating diverse functions such as growth, metabolism, and immune responses.
Protein-Ligand Interactions: Molecular Recognition
Proteins can also interact with small molecules called ligands. Receptor proteins, for instance, bind to ligands such as hormones, neurotransmitters, or drugs, initiating cellular responses. These interactions rely on specific binding sites and molecular recognition.
Protein-DNA Interactions: Controlling Genetic Information
Transcription factors, a class of proteins, interact with DNA to regulate gene expression. They bind to specific DNA sequences, promoting or inhibiting transcription, thereby controlling RNA and protein synthesis.
Membrane Proteins: Regulating Cellular Transport
Integral membrane proteins participate in macromolecular interactions by regulating the transport of ions and molecules across cell membranes. Transport proteins, ion channels, and pumps interact precisely to maintain cellular homeostasis.
Cooperativity and Allosteric Regulation: Fine-Tuning Cellular Processes
Cooperativity and allosteric regulation are mechanisms that modulate protein function. In cooperativity, binding one ligand to a protein influences the binding of subsequent ligands, often amplifying the response. Allosteric regulation occurs when a molecule binds to a site other than the active site, altering the protein's conformation and activity.
Interactions in Signaling Pathways: Cellular Communication
Signal transduction pathways rely on cascades of macromolecular interactions to transmit extracellular signals into cellular responses. Kinases and phosphatases, enzymes that add or remove phosphate groups, play pivotal roles in these pathways.
Protein Folding and Misfolding: Disease Implications
Proteins must fold into specific three-dimensional shapes to function correctly. Misfolded proteins can lead to Alzheimer's, Parkinson's, and prion diseases. Chaperone proteins assist in proper protein folding and prevent aggregation.
References
Voet, D., Voet, J. G., & Pratt, C. W. (2016). Fundamentals of Biochemistry: Life at the Molecular Level. Wiley.
Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2017). Lehninger Principles of Biochemistry. W. H. Freeman.
Berg, J. M., Tymoczko, J. L., & Stryer, L. (2002). Biochemistry. W. H. Freeman
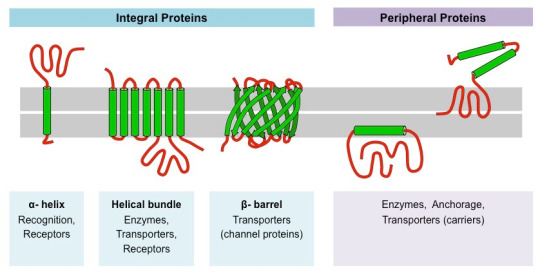
#science#college#biology#education#school#medicine#student#doctors#health#healthcare#proteins#molecular biology#molecular structure#chemestry#chemistry
98 notes
·
View notes
Text
molecular geometry my sweetheart my baby. electron orbitals kill yourself.
#I am slowly and steadily losing the plot#chemistry#chemistry meme#chemistry memes#a level chemistry meme#a level chemistry#molecular geometry#molecular structure#molecules#molecular memes#electron configuration#electron geometry#VSERP#atom memes#is this physics?#fuck it#physics meme
46 notes
·
View notes
Text
I’m dumb but convinced this says something, does anyone know how to read molecular structures?? Got this at the thrift shop yesterday.

12 notes
·
View notes
Video
Hey! by Katsuaki Shoda Via Flickr: Canon EOS R6m2 + RF24-105mm F4L IS USM
#Kobe#Hyogo#Japan#No People#Organic#Molecule#Abstract#Molecular Structure#Atom#Sign#Hand Sign#Sign Language#Figurine#Iron - Metal#flickr
2 notes
·
View notes
Text
Weather resistance and corrosion resistance of PVDF
In the vast starry sky of material science, polyvinylidene fluoride (PVDF) is like a shining star, illuminating the way for many industrial applications with its excellent weather resistance and corrosion resistance. As a high-performance fluoropolymer, PVDF stands out among many materials with its unique molecular structure and chemical properties, and has become an indispensable key material in…
#chemical equipment#construction field#corrosion resistance#Electrical Insulation#fluorine atom#molecular structure#PVDF#weather resistance
0 notes
Link
All our links in one place:https://sleekbio.com/aanews69Visit our Patreonhttps://www.patreon.com/DNPLServicesWe deliver stories. We also give you guides, tip...
#water molecule#polar molecule#chemistry#science education#science explainer#hydrogen bonds#h2o#molecular structure#chemistry concepts#water polarity#educational#atom#electron#oxygen atom#chemical bonding#dipole moment#partial charges#high school science#university level science#basic chemistry.
1 note
·
View note
Text
In the next several sections, we describe how these measurements are made for nuclear spin states of these two isotopes and then how this information can be correlated with molecular structure.
"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quote#chemistry#nonfiction#textbook#measurement#nuclear spin#isotope#carbon 13#hydrogen 1#carbon#hydrogen#molecular structure
0 notes
Text
(part 4) fun for the whole family continues!!















did you spot rumble? :)
next
prev
start
#teenagers man#can't live with em can't live without em#HELLO AGAIN...I WILL TRY TO UPDATE EVERY SUNDAY!!#every time i post a new part i get so nervous HGIHGKLH my heart is beating so fast LMAOOO#who's soundwave tryin to convince? frenzy or himself? up to interpretation...#OK IM GLAD EVEYRONE'S BEEN ENJOYING THE LONG FORMAT!! it's much easier to draw and plot out for me as well!#the next part is 20 pages. molecular structure collapses and i scatter into the wind#DON'T DO THIS DAD#transformers#maccadam#soundwave#frenzy#transformers au#tf art#artists on tumblr#tf au#humanformers#ive seriously been waking up thinking about this au and goign to sleep thinking about this au. someone get me out of here
1K notes
·
View notes
Text

(LATE) MAID DAY!!!!!!!!!
extra:



#null rot#Hantengu#hantengu clones#Sekido#Karaku#Urogi#Aizetsu#kny#kimetsu no yaiba#demon slayer#メイドの日#maid day#I FUCKING LOVE MAIDS AND BUTLERS#GREAT GREAT GREAT#I HAD THIS PLANNED IN ADVANCE BUT I MISSED THE ACTUAL DAY GRGRHHGHGGHHH#anyway. this was just another excuse to dress them up. i love using these idiots as my dress up dolls#shakes you aggressively. can you see the common theme whenever i dress them??#for the end bit. sekido was also embarrassed. while nothing ACTUALLY showed. he was still technically upskirted#karaku is surprised to be the one upskirted this time. Aizetsu is sad because this couldve ended badly#and urogi—like sekido—didnt have anything shown bc it was too frilly but is still embarrassed bc he was technically upskirted too#love them all. buries you into the ground. love them!!!#sorry for always making karaku pose like a diva. i saw him walk with his hand on his hip once and it changed my molecular structure
425 notes
·
View notes
Text

murderous tendencies be damned my boy can work a violin
#aka finished that erik wip today#you may ask. kaya why do you use the same color pallet for everything. and i will reply with: because i like it and i can#are the squiggles sound or are they secondary structure proteins the world will never know#this is what i get for coloring during cell and molecular bio#phantom of the opera#poto#erik phantom of the opera#erik poto#erik the phantom#phantom of the opera art#my art tag
2K notes
·
View notes
Text


You are what you eat and baby I’m always making that jello jigglers resippy 😏
#has anyone ever compared the two recipes tho#like what does the addition of cold water do to the molecular structure of jello can I get gay molecular chemistry tumblr on this#gpoy#gpoyb
2K notes
·
View notes
Text
You ever have arguably too much Nutella and it like fucks you up a little
#something happened to my atoms#my molecular structure is unstable#its pancake tuesday i wrnt too hard
132 notes
·
View notes
Text
Atom Eve next season needs less time as Mark’s therapist and more time going balls to wall w/ her OP powers!!!! I’m tired of this green lantern object generation, have her turn the air in a viltrumite’s lungs into mayonnaise!!!!!!!
#this woman could forge a dwarf star in her hands and collapse it into a black hole#and you keep making her generate pink walls that don’t even work#GOD#if you didnt want her overpowered THEN DONT MAKE HER POWER THAT CRAZY#firestorm has her powers from what i’ve seen of him in jl action#but he needs to know the molecular structure!#EVE COULD’VE TURNED CONQUEST’S CLOTHES INTO DARK MATTER AND IMPLODED HIM#invincible#still loved s3#atom eve
22 notes
·
View notes
Text
My brain, every other second for the past 6 weeks straight gay: what if we rewatched Agatha All Along?
#i'm not even kidding that gay ass witch show has rewritten my molecular structure#agatha all along#agatha harkness#i just really like witchy stuff and women and gay shenannigans like i was an easy target for them
31 notes
·
View notes
Text


what's stronger than a diamond?
take this antarctic empire inspired fusion design of phil and techno! he was real fun to design (my favourite bit is his cape, it turns into mist at the very bottom like a waterfall (or into snow, your choice)) :D
also fun fact, this was lined with a feather pen that i had to dip into an inkwell :D my lining pen was broken by my siblings but this had the perfect nib !
red king design by @cherrifire + the gemcyt au by @chrisrin!
stupid doodle under the cut:

#he gets to swear a bit#because phil swears like a sailor and techno doesn't so it evens out#he gets to say what the hell#the hexagon around his eye is a reference to how lonsdaleite is stronger than diamond due to its hexagonal molecular structure!#(versus the cube structure diamond has)#philza#technoblade#rendog#renthedog#???#martyn inthelittlewood#inthelittlewood#gemcyt#also the idea of ren and techno being rivals has been funny to me since 3rd life#they have never interacted but the comparisons drawn between them were SO FUNNY#enderinks
250 notes
·
View notes
Text

First little group of intros, more coming soon. :)





#homestuck#ask blog#send asks#ask#ask me anything#anon ask#intro post#blog intro#pinned intro#homestuck oc#fankids#fanchild#au#fanchild au#my au#ocs#oc art#oc intro#gay#seawed joe molecular structure vantas-ampora#julius captor#blake ampora#shelle ampora#men#boy kissing#part1#wip#wap#pride
10 notes
·
View notes