#p53 mutation
Text
youtube
#Endometrial cancer#Gynecologic oncology#Clinical trials#Immunotherapy#Targeted therapies#Molecular classification#Precision medicine#Cancer biomarkers#GCIG#p53 mutation#Microsatellite instability-high#Checkpoint inhibitors#Oncology consensus#Therapeutic advancements#Patient stratification#Standardized outcomes#Collaborative research#PTEN mutation#PIK3CA mutation#Recurrent cancer.#Youtube
0 notes
Text
De-extinction startup Colossal Biosciences wants to bring back the woolly mammoth. Well, not the woolly mammoth exactly, but an Asian elephant gene-edited to give it the fuzzy hair and layer of blubber that allowed its close relative to thrive in sub-zero environments.
To get to these so-called “functional mammoths,” Colossal’s scientists need to solve a whole bunch of challenges: making the right genetic tweaks, growing edited cells into fully formed baby functional mammoths, and finding a space where these animals can thrive. It’s a long, uncertain road, but the startup has just announced a small breakthrough that should ease some of the way forward.
Scientists at Colossal have managed to reprogram Asian elephant cells into an embryonic-like state that can give rise to every other cell type. This opens up a path to creating elephant sperm and eggs in the lab and being able to test gene edits without having to frequently take tissue samples from living elephants. The research, which hasn’t yet been released in a peer-reviewed scientific journal, will be published on the preprint server Biorxiv.
There are only around 30,000 to 50,000 Asian elephants in the wild, so access to these animals—and particularly their sperm and eggs—is extremely limited. Yet Colossal needs these cells if they’re going to figure out how to bring their functional mammoths to life. “With so few fertile female elephants, we really don’t want to interfere with their reproduction at all. We want to do it independently,” says George Church, a Harvard geneticist and Colossal cofounder.
The cells that Colossal created are called induced pluripotent stem cells (iPSCs), and they behave a lot like the stems cells found in an embryo. Embryonic stem cells have the ability to give rise to all kinds of different cell types that make up organisms—a quality that scientists call pluripotency. Most cells, however, lose this ability as the organism develops. Human skin, for instance, can’t spontaneously turn into muscle or cells that line the inside of the intestine.
In 2006, the Japanese scientist Shinya Yamanaka showed it was possible to take mature cells and turn them back into a pluripotent state. Yamanaka’s research was in mice cells, but later scientists followed up by deriving iPSCs for lots of different species, including humans, horses, pigs, cattle, monkeys, and the northern white rhino—a functionally extinct subspecies with only two individuals, both females, remaining in the wild.
Reprogramming Asian elephant cells into iPSCs proved trickier than with other species, says Eriona Hysolli, head of biological sciences at Colossal. As with other species, the scientists reprogrammed the elephant cells by exposing them to a series of different chemicals and then adding proteins called transcription factors that turn on particular genes to change how the cells functions. The whole process took two months, which is much longer than the 5 to 10 days it takes to create mouse iPSCs or the three weeks for human iPSCs.
This difficulty might have to do with the unique biology of elephants, says Vincent Lynch, a developmental biologist at the University at Buffalo in New York who wasn’t involved in the Colossal study. Elephants are the classic example of Peto’s paradox—the idea that very large animals have unusually low rates of cancer given their size. Since cancer can be caused by genetic mutations that accumulate as cells divide, you’d expect that animals with 100 times more cells than humans would have a much higher risk of cancer.
But elephants have cancer rates even lower than humans—a surprising fact given their vast size. One hypothesis for elephants’ cancer-defying biology is that they carry lots of copies of a tumor-suppressing gene called P53. Humans, on the other hand, only have one copy of this gene.
P53 is good for elephant health, but it could be the reason that up until now scientists have struggled to create iPSCs from elephant cells, Lynch says. One way the gene seems to work is by stopping cells from entering a state where they can duplicate indefinitely, which is one of the key features of iPSCs.
Hysolli says that she’d like to reduce the time it takes to create elephant iPSCs, and refine the process so the Colossal team can produce them at a greater scale. The iPSCs will be particularly useful if Colossal’s scientists can turn them into sperm and egg cells, something that Hysolli’s team is already working on. Since there is a relatively limited supply of elephant eggs and sperm, one problem facing the de-extinction project is getting enough genetic diversity to support a population of functional mammoths—develop them from too few individuals, and you risk the negative effects of inbreeding. Being able to create sperm and egg cells in the lab should help with that, Church says.
These cells could also be useful for conservation work, Hysolli says. Colossal has partnered with researchers working on elephant endotheliotropic herpes virus (EEHV), a leading cause of death for young Asian elephants. The iPSCs could be a good way to figure out how the virus infects different cell types. The cells will also be useful for testing whether Colossal’s edits to produce mammoth-like fur and fat layers are working as scientists hope.
“I have no doubt that given enough time and money they will overcome the technical challenges of making a woolly-mammoth-looking elephant,” says Lynch. But he’s less convinced of the ecological benefits of de-extinction. The startup intends to introduce the elephant-mammoth hybrids into the wild to re-create the role once played by the mammoth in the Arctic ecosystem, grazing the land and trampling snow cover, potentially decelerating the melting of permafrost.
“How many hairy Asian elephants do you need to make that work?” Lynch asks. Whether there really is a niche for edited elephants in the Arctic 4,000 years after mammoths last roamed the area is a question that conservationists are still grappling with. Sure, scientists might be able to create mammoth-like Asian elephants, but whether we should is open to much debate.
Colossal’s scientists will be glad if they get to that point. Although they have elephant iPSCs, much of the work of creating elephant-mammoth hybrids is ahead of them. They must figure out how to create elephant sperm and egg cells, master the right edits to tweak their elephants, and take their creation through the 22-month Asian elephant gestation period. And then they have to do it enough times to build a population that can actually deliver on some of their ecological aims.
“It feels very significant,” Church says of the iPSC breakthrough. “This is a very big deal.” If Colossal is going to deliver on its de-extinction mission, then there will be many other moments like this ahead.
6 notes
·
View notes
Note
CJ, throwing you the biggest distraction I can think of. Please explain to me p53 role in cancer 😭 I watch signaling pathway everything!!! Go!!
The TP53 gene is a tumour suppressor gene so it stops the development of tumours. If a person only inherits one functional copy of the gene, their risk of developing multiple cancers in their lifetime is higher (Li-Fraumeni syndrome, it’s rare). More commonly, mutations in the gene are what lead to the development of cancers.
This gene is found on chromosome 17 and provides instructions to make the protein P53. This protein remains in the nucleus of cells and it binds directly to DNA. When DNA becomes damaged, P53 plays a role in determining whether the damaged DNA can be repaired. If it can be repaired, the proper genes will be activated to fix it. If it can’t, P53 will prevent the DNA from undergoing replication and trigger apoptosis (programmed cell death). This stops the damaged DNA from mutating and dividing further.
If P53 is damaged, missing, or suppressed, there is a chance this tumour checkpoint can be missed. In that case, it is much more likely that damaged DNA can “get through” and pass through replication and transcription into RNA and further mutate into cancer.
2 notes
·
View notes
Text
Tumor Suppressor p53 Protein , Victor McKusick, Mendelian Inheritance in Man, 1966.
Here I present: “Tumor Suppressor p53 Protein”, Victor McKusick, Mendelian Inheritance in Man’, 1966.
INTRODUCTION.
Tumor Suppressor p53 Protein encoded by the TP53 gene, is the most frequently mutated gene in cancer.
There is evidence that the tumor protein p53-binding protein-2 (TP53BP2) gene is on cytogenetic location 1q41 and genomic coordinates1:223,779,893-223,845,947 . The screenshot of…
0 notes
Text
Patrick Moscatiello: A Legacy of Hope and Advocacy, told by his brother Dan Moscatiello

"Take charge of your health, be your own advocate—it could save your life." This was the mantra Patrick Moscatiello lived by, which shaped his remarkable journey through life and multiple life-threatening cancer diagnoses. Diagnosed with jawbone osteosarcoma at just 24, Patrick faced a daunting prognosis that gave him a 50-50 chance of survival; remarkably, he beat it. At the age of 33, genetic testing revealed a disorder—a genetic mutation of the P53 gene called Li-Fraumeni Syndrome (LFS)—which would paradoxically change his life for the better. His spirit was strengthened, and he became a better version of himself, despite having every reason to falter.
Patrick consistently maintained the right mindset and energy to overcome every obstacle in his life. He was that guy that you wanted to talk to, he inspired everyone. His dedication to advocating for genetic awareness and proactive health measures significantly benefited thousands in the LFS community through his close work with the LFS Association. At age 41, he was diagnosed with a medulloblastoma brain tumor that was removed in July 2020, diagnosed with drop metastasis discovered during Christmas of 2021, and ultimately led to leptomeningeal disease April 2022; he tragically passed away on June 5, 2022. During those two years, we lived through our worst experiences alongside him.
Patrick transformed not only his own fate but also influenced the outlook of those around him on the importance of being your own health advocate. His story is not just about the battles he fought; it's a call to action, inspiring all of us to take charge of our health and understand the power of our choices. Learn from him: despite passing at 41, Patrick led an impactful life with his amazing partner, Christina Moscatiello, and a loving family. He was the last living male in the Moscatiello family to carry the genetic disorder of LFS, and he was determined to ensure it ended with him.
How It All Started: A Game-Changing Moment
Nobody ever thinks a pool volleyball game will ever change your life but that's exactly what Patrick would experience. In a fleeting moment filled with laughter and light-hearted competition, a slip on the wet deck altered the course of his life forever. He jumped out of the pool to retrieve the ball, but as he ran, his feet lost grip on the wet concrete surface. Patrick slipped and fell, landing chin-first on the concrete—a minor accident that unbeknownst to anyone, would have major consequences. At first glance, the injury seemed trivial but the impact had a far deeper effect than anyone could initially see. I visited him just a few days later, and he showed me a lump bulging from his bottom gums and teeth: a large, abnormal mass had formed at the bottom of his jaw only observed when opening his mouth. Concern was evident in his eyes—something was wrong, though neither of us could have imagined the gravity of it at that moment nor how to handle it.
We, naive and uninformed, tried to downplay the severity, making a dentist appointment hoping it was nothing more to worry about. Yet, deep down, there was a scary sense of something seriously wrong. It was during a follow-up visit with an orthopedic surgeon that our fears were confirmed. The surgeon took one look and his attention and concern scared Patrick into action, he performed a biopsy, and the results were as dire as they come: it was cancer.
The diagnosis was not just a shock; it was the nightmares that echoed through our family history. Patrick was faced with osteosarcoma—the same malignant force that had claimed our uncle Patrick at a young age. Suddenly, the chilling pattern of our family's battles with cancer was impossible to ignore. Patrick's life was upended in an instant; he left his home, his belongings, and his old life behind to fight this new battle home with his family. He underwent treatments at Fox Chase, where he was given a stark prognosis—a 50/50 chance to survive and a 24 month ordeal. Patrick not only survived but emerged from his ordeal fundamentally transformed, living his subsequent years as a better version of himself. The entire experience, while deeply traumatizing and devastating, served as a profound turning point not just for him but for our entire family. We would never be the same, we found a way to be better because of Patrick transforming behavior through the most challenging times.
The “Curse” of Cancer and the Clarity of Genetic Testing
Growing up, the shadow of cancer loomed over our family, a haunting legacy that deeply affected my father and his 2 remaining brothers. He endured the heartbreak of losing his older sister, Jane, to brain cancer at just six years old, and his younger brother, Patrick (our uncle), to bone cancer, this started at a young age and he would pass at fifteen. His mother succumbed to breast cancer at the age of 55, just after learning her hope that my father would have a daughter, she passed after Lisa’s birth. These losses lacked any medical understanding and were simply coined a “curse” by an older generation without the medical resources that exist today. Certainly my fathers experiences were traumatic and with so much unknown left fear of reliving such loss in the next generation. My father was forever changed by these tragedies, carrying a burden of grief that would resurface with his own son's cancer.
This was finally understood when Patrick, after his battle with jawbone cancer in 2005 that nearly claimed his life, underwent genetic testing nine years later. The results confirmed Li-Fraumeni Syndrome (LFS), a rare genetic condition that predisposed him to cancer, echoing the fears and struggles that had “cursed” our family. The revelation forced my father to relive his worst nightmares, his childhood trauma revisited upon his second son, aging him beyond his years in a way words can hardly describe. My mother, equally pained yet unwavering, bolstered Patrick’s bravery with her love and support. Patrick would always credit our mother for allowing him to be brave.
Patrick rejected this idea that we were “cursed”. He responded to any mention of a “curse”, often saying, “How can I be cursed when I am so loved?” That was Patrick’s energy and mindset and the euphoria he brought into every room that profoundly influenced everyone around him. This shift in perspective ultimately transformed our approach to healthcare and deepened our understanding of our family’s past, guiding us toward a future where we would not look at any diagnosis in despair but love your fate and who you are. He can find something good in the worst of it and he never seemed to disappoint in that. Patrick changed the entire Moscatiello family mindset and transformed our despair into proactive action.
Advocate for Change
Now educated and understanding the underlying risk with his genetic disorder, he took this information and changed the narrative in our family. We would have a new perspective on his condition, be proactive, get ahead of your health as proactive care can reduce your risk. Everyone one previously in the dark about our history was now empowered to know if they stand a similar fate. Patrick became an advocate for genetic testing. He educated others about its importance, breaking down the barriers of fear and uncertainty that often surround such discussions. Patrick’s efforts were not just about prevention but empowerment—encouraging individuals and families to take proactive steps in their healthcare. If you are reading this and genetic testing was suggested to you, go get it done! Anyone who has a loss from cancer, this story is more relevant to you than you think.
Enduring Impact
In the final years of his life, Patrick faced one of the worst diagnoses with incredible courage and dignity. He taught us how to live and how to die. We could see that Patrick lived longer in 41 years than most people that live more than 80 years. He spoke openly about what is most important in life, influencing countless lives through his incredible talent to positively influence how one thought about a particular subject, helping bring clarity to what is important and what is not. His words, filled with humor and hope, resonated deeply with those who heard them, leaving a lasting impression that would outlive his physical presence. We love him forever.
The impact of Patrick’s presence and spirit are difficult to capture in words. A passage from his journal beautifully illustrates the essence of his character: 'To spend each day as if it were my last, without frenzy, laziness, or any pretending. To love life, even in darkness and to influence others to do the same.' Patrick had a unique way of viewing life’s challenges. While many might despair under the weight of a figurative piano hanging overhead, asking 'Why me?', Patrick’s perception was, 'Why not me?'. He embraced the possibility of change, understanding that this freedom to transform was key to becoming the better version of himself. He changed the idea of a perceived 'curse' into a 'gift.' His outlook never wavered; he learned to find joy in every circumstance, which in turn enriched his life immensely. This shift redefined his—and consequently our—understanding of true wealth and power. No longer measured in dollars and cents, but in health and happiness. Patrick’s message to us all is clear: Choose happiness, be grateful, embrace your destiny, and love who you are. Every situation holds a some goodness; it only requires the right mindset to see it. Such was the profound energy Patrick projected to everyone around him.
A Brother’s Reflection
Patrick's journey was more than a series of cancer battles; it was a profound way he viewed life, love, and his human spirit. His legacy is not marked by his disease but by the lives he touched and the positive changes he inspired. As we remember Patrick, we celebrate not just his life but the spirit that he nurtured in all of us. I feel his energy today, we never forget him, we will never get over him, he deserves that. I have learned, and others close to him, that we will never get over losing him. We are learning that it's ok to carry the pain of his loss, it's a way to thank him for everything he has done for us. I am so lucky to have a brother like Patrick.
For anyone needing guidance, remember Patrick's approach: Control your perceptions, direct your actions wisely, and accept what's beyond your control. These principles alone can profoundly shape your life.
0 notes
Link
0 notes
Photo

Using adavosertib-encapsulated MOFs for p53-mutated gallbladder cancer treatment via synthetic lethality
0 notes
Text
Scientists develop a rapid gene-editing screen to find effects of cancer mutations
New Post has been published on https://sunalei.org/news/scientists-develop-a-rapid-gene-editing-screen-to-find-effects-of-cancer-mutations/
Scientists develop a rapid gene-editing screen to find effects of cancer mutations

Tumors can carry mutations in hundreds of different genes, and each of those genes may be mutated in different ways — some mutations simply replace one DNA nucleotide with another, while others insert or delete larger sections of DNA.
Until now, there has been no way to quickly and easily screen each of those mutations in their natural setting to see what role they may play in the development, progression, and treatment response of a tumor. Using a variant of CRISPR genome-editing known as prime editing, MIT researchers have now come up with a way to screen those mutations much more easily.
The researchers demonstrated their technique by screening cells with more than 1,000 different mutations of the tumor suppressor gene p53, all of which have been seen in cancer patients. This method, which is easier and faster than any existing approach, and edits the genome rather than introducing an artificial version of the mutant gene, revealed that some p53 mutations are more harmful than previously thought.
This technique could also be applied to many other cancer genes, the researchers say, and could eventually be used for precision medicine, to determine how an individual patient’s tumor will respond to a particular treatment.
“In one experiment, you can generate thousands of genotypes that are seen in cancer patients, and immediately test whether one or more of those genotypes are sensitive or resistant to any type of therapy that you’re interested in using,” says Francisco Sanchez-Rivera, an MIT assistant professor of biology, a member of the Koch Institute for Integrative Cancer Research, and the senior author of the study.
MIT graduate student Samuel Gould is the lead author of the paper, which appears today in Nature Biotechnology.
Editing cells
The new technique builds on research that Sanchez-Rivera began 10 years ago as an MIT graduate student. At that time, working with Tyler Jacks, the David H. Koch Professor of Biology, and then-postdoc Thales Papagiannakopoulos, Sanchez-Rivera developed a way to use CRISPR genome-editing to introduce into mice genetic mutations linked to lung cancer.
In that study, the researchers showed that they could delete genes that are often lost in lung tumor cells, and the resulting tumors were similar to naturally arising tumors with those mutations. However, this technique did not allow for the creation of point mutations (substitutions of one nucleotide for another) or insertions.
“While some cancer patients have deletions in certain genes, the vast majority of mutations that cancer patients have in their tumors also include point mutations or small insertions,” Sanchez-Rivera says.
Since then, David Liu, a professor in the Harvard University Department of Chemistry and Chemical Biology and a core institute member of the Broad Institute, has developed new CRISPR-based genome editing technologies that can generate additional types of mutations more easily. With base editing, developed in 2016, researchers can engineer point mutations, but not all possible point mutations. In 2019, Liu, who is also an author of the Nature Biotechnology study, developed a technique called prime editing, which enables any kind of point mutation to be introduced, as well as insertions and deletions.
“Prime editing in theory solves one of the major challenges with earlier forms of CRISPR-based editing, which is that it allows you to engineer virtually any type of mutation,” Sanchez-Rivera says.
When they began working on this project, Sanchez-Rivera and Gould calculated that if performed successfully, prime editing could be used to generate more than 99 percent of all small mutations seen in cancer patients.
However, to achieve that, they needed to find a way to optimize the editing efficiency of the CRISPR-based system. The prime editing guide RNAs (pegRNAs) used to direct CRISPR enzymes to cut the genome in certain spots have varying levels of efficiency, which leads to “noise” in the data from pegRNAs that simply aren’t generating the correct target mutation. The MIT team devised a way to reduce that noise by using synthetic target sites to help them calculate how efficiently each guide RNA that they tested was working.
“We can design multiple prime-editing guide RNAs with different design properties, and then we get an empirical measurement of how efficient each of those pegRNAs is. It tells us what percentage of the time each pegRNA is actually introducing the correct edit,” Gould says.
Analyzing mutations
The researchers demonstrated their technique using p53, a gene that is mutated in more than half of all cancer patients. From a dataset that includes sequencing information from more than 40,000 patients, the researchers identified more than 1,000 different mutations that can occur in p53.
“We wanted to focus on p53 because it’s the most commonly mutated gene in human cancers, but only the most frequent variants in p53 have really been deeply studied. There are many variants in p53 that remain understudied,” Gould says.
Using their new method, the researchers introduced p53 mutations in human lung adenocarcinoma cells, then measured the survival rates of these cells, allowing them to determine each mutation’s effect on cell fitness.
Among their findings, they showed that some p53 mutations promoted cell growth more than had been previously thought. These mutations, which prevent the p53 protein from forming a tetramer — an assembly of four p53 proteins — had been studied before, using a technique that involves inserting artificial copies of a mutated p53 gene into a cell.
Those studies found that these mutations did not confer any survival advantage to cancer cells. However, when the MIT team introduced those same mutations using the new prime editing technique, they found that the mutation prevented the tetramer from forming, allowing the cells to survive. Based on the studies done using overexpression of artificial p53 DNA, those mutations would have been classified as benign, while the new work shows that under more natural circumstances, they are not.
“This is a case where you could only observe these variant-induced phenotypes if you’re engineering the variants in their natural context and not with these more artificial systems,” Gould says. “This is just one example, but it speaks to a broader principle that we’re going to be able to access novel biology using these new genome-editing technologies.”
Because it is difficult to reactivate tumor suppressor genes, there are few drugs that target p53, but the researchers now plan to investigate mutations found in other cancer-linked genes, in hopes of discovering potential cancer therapies that could target those mutations. They also hope that the technique could one day enable personalized approaches to treating tumors.
“With the advent of sequencing technologies in the clinic, we’ll be able to use this genetic information to tailor therapies for patients suffering from tumors that have a defined genetic makeup,” Sanchez-Rivera says. “This approach based on prime editing has the potential to change everything.”
The research was funded, in part, by the National Institute of General Medical Sciences, an MIT School of Science Fellowship in Cancer Research, a Howard Hughes Medical Institute Hanna Gray Fellowship, the V Foundation for Cancer Research, a National Cancer Institute Cancer Center Support Grant, the Ludwig Center at MIT, a Koch Institute Frontier Award, the MIT Research Support Committee, and the Koch Institute Support (core) Grant from the National Cancer Institute.
0 notes
Text
Scientists develop a rapid gene-editing screen to find effects of cancer mutations
New Post has been published on https://thedigitalinsider.com/scientists-develop-a-rapid-gene-editing-screen-to-find-effects-of-cancer-mutations/
Scientists develop a rapid gene-editing screen to find effects of cancer mutations


Tumors can carry mutations in hundreds of different genes, and each of those genes may be mutated in different ways — some mutations simply replace one DNA nucleotide with another, while others insert or delete larger sections of DNA.
Until now, there has been no way to quickly and easily screen each of those mutations in their natural setting to see what role they may play in the development, progression, and treatment response of a tumor. Using a variant of CRISPR genome-editing known as prime editing, MIT researchers have now come up with a way to screen those mutations much more easily.
The researchers demonstrated their technique by screening cells with more than 1,000 different mutations of the tumor suppressor gene p53, all of which have been seen in cancer patients. This method, which is easier and faster than any existing approach, and edits the genome rather than introducing an artificial version of the mutant gene, revealed that some p53 mutations are more harmful than previously thought.
This technique could also be applied to many other cancer genes, the researchers say, and could eventually be used for precision medicine, to determine how an individual patient’s tumor will respond to a particular treatment.
“In one experiment, you can generate thousands of genotypes that are seen in cancer patients, and immediately test whether one or more of those genotypes are sensitive or resistant to any type of therapy that you’re interested in using,” says Francisco Sanchez-Rivera, an MIT assistant professor of biology, a member of the Koch Institute for Integrative Cancer Research, and the senior author of the study.
MIT graduate student Samuel Gould is the lead author of the paper, which appears today in Nature Biotechnology.
Editing cells
The new technique builds on research that Sanchez-Rivera began 10 years ago as an MIT graduate student. At that time, working with Tyler Jacks, the David H. Koch Professor of Biology, and then-postdoc Thales Papagiannakopoulos, Sanchez-Rivera developed a way to use CRISPR genome-editing to introduce into mice genetic mutations linked to lung cancer.
In that study, the researchers showed that they could delete genes that are often lost in lung tumor cells, and the resulting tumors were similar to naturally arising tumors with those mutations. However, this technique did not allow for the creation of point mutations (substitutions of one nucleotide for another) or insertions.
“While some cancer patients have deletions in certain genes, the vast majority of mutations that cancer patients have in their tumors also include point mutations or small insertions,” Sanchez-Rivera says.
Since then, David Liu, a professor in the Harvard University Department of Chemistry and Chemical Biology and a core institute member of the Broad Institute, has developed new CRISPR-based genome editing technologies that can generate additional types of mutations more easily. With base editing, developed in 2016, researchers can engineer point mutations, but not all possible point mutations. In 2019, Liu, who is also an author of the Nature Biotechnology study, developed a technique called prime editing, which enables any kind of point mutation to be introduced, as well as insertions and deletions.
“Prime editing in theory solves one of the major challenges with earlier forms of CRISPR-based editing, which is that it allows you to engineer virtually any type of mutation,” Sanchez-Rivera says.
When they began working on this project, Sanchez-Rivera and Gould calculated that if performed successfully, prime editing could be used to generate more than 99 percent of all small mutations seen in cancer patients.
However, to achieve that, they needed to find a way to optimize the editing efficiency of the CRISPR-based system. The prime editing guide RNAs (pegRNAs) used to direct CRISPR enzymes to cut the genome in certain spots have varying levels of efficiency, which leads to “noise” in the data from pegRNAs that simply aren’t generating the correct target mutation. The MIT team devised a way to reduce that noise by using synthetic target sites to help them calculate how efficiently each guide RNA that they tested was working.
“We can design multiple prime-editing guide RNAs with different design properties, and then we get an empirical measurement of how efficient each of those pegRNAs is. It tells us what percentage of the time each pegRNA is actually introducing the correct edit,” Gould says.
Analyzing mutations
The researchers demonstrated their technique using p53, a gene that is mutated in more than half of all cancer patients. From a dataset that includes sequencing information from more than 40,000 patients, the researchers identified more than 1,000 different mutations that can occur in p53.
“We wanted to focus on p53 because it’s the most commonly mutated gene in human cancers, but only the most frequent variants in p53 have really been deeply studied. There are many variants in p53 that remain understudied,” Gould says.
Using their new method, the researchers introduced p53 mutations in human lung adenocarcinoma cells, then measured the survival rates of these cells, allowing them to determine each mutation’s effect on cell fitness.
Among their findings, they showed that some p53 mutations promoted cell growth more than had been previously thought. These mutations, which prevent the p53 protein from forming a tetramer — an assembly of four p53 proteins — had been studied before, using a technique that involves inserting artificial copies of a mutated p53 gene into a cell.
Those studies found that these mutations did not confer any survival advantage to cancer cells. However, when the MIT team introduced those same mutations using the new prime editing technique, they found that the mutation prevented the tetramer from forming, allowing the cells to survive. Based on the studies done using overexpression of artificial p53 DNA, those mutations would have been classified as benign, while the new work shows that under more natural circumstances, they are not.
“This is a case where you could only observe these variant-induced phenotypes if you’re engineering the variants in their natural context and not with these more artificial systems,” Gould says. “This is just one example, but it speaks to a broader principle that we’re going to be able to access novel biology using these new genome-editing technologies.”
Because it is difficult to reactivate tumor suppressor genes, there are few drugs that target p53, but the researchers now plan to investigate mutations found in other cancer-linked genes, in hopes of discovering potential cancer therapies that could target those mutations. They also hope that the technique could one day enable personalized approaches to treating tumors.
“With the advent of sequencing technologies in the clinic, we’ll be able to use this genetic information to tailor therapies for patients suffering from tumors that have a defined genetic makeup,” Sanchez-Rivera says. “This approach based on prime editing has the potential to change everything.”
The research was funded, in part, by the National Institute of General Medical Sciences, an MIT School of Science Fellowship in Cancer Research, a Howard Hughes Medical Institute Hanna Gray Fellowship, the V Foundation for Cancer Research, a National Cancer Institute Cancer Center Support Grant, the Ludwig Center at MIT, a Koch Institute Frontier Award, the MIT Research Support Committee, and the Koch Institute Support (core) Grant from the National Cancer Institute.
#000#Adenocarcinoma#approach#artificial#Biology#biotechnology#Broad Institute#Cancer#cancer cells#cell#Cells#change#chemical#chemistry#CRISPR#data#delete#Design#development#DNA#drugs#Editing#Editing Technique#effects#efficiency#Engineer#engineering#enzymes#Forms#Foundation
0 notes
Text
Is cancer hereditary?
Is cancer hereditary?
Yes, some cancers are genetic or hereditary. They are seen more often in successive generations. A person with a family history of cancer has a higher risk of developing cancer.
What you need to know if more than one family member has contracted the cancer is to find out what type of cancer your family member (s) had and whether it is related to the risk of the family inheritance. Cancer is caused by gene effects and can be passed on from generation to generation, but the probability of it occurring in any blood ratio is not very high. There are also cancers that are not genetic or familial. Other factors such as eating habits, lifestyle such as smoking, and socioeconomic status can affect it.
Cancer due to Gene Mutation:
About 3% to 10% of all cancers are genetic or hereditary. It is the result of a genetic mutation that is passed on from generation to generation. Cancers such as breast cancer, ovarian cancer, colon cancer, and uterine cancer are not commonly inherited, but people with a family history of cancer have a higher risk of developing cancer than others.
Does every gene mutation cause cancer?
Not every genetic defect results in cancer. The genes whose defects cause cancer are called cancer susceptibility genes. People having mutations known as BRCA or p53 are especially prone to cancers. These gene mutations are tested among healthy family members by cancer specialists by genetic screening.
Keep in mind that hereditary cancers and genetic cancers are not always the same although these terms appear to be synonymous. Most cancers have a genetic basis, due to a genetic mutation that is transmitted later in life.
What to do if any blood relative has had cancer?
If in doubt, consult a cancer specialist in your area. These doubts may arise if more than one family member has the same type of cancer or if the family has had cancer caused by the same genetic mutation (for example breast and ovarian cancer in different family members).
If your oncologist suggests that you may have a genetic mutation after studying your family history, he or she may ask you to undergo genetic screening.
0 notes
Text
Unusual rigour – the GRG Health difference in Research
Sometimes, a new client wants to know what makes GRG Health different. So, GRG Health sends out a suitable presentation which satisfies clients at least 80% of the time.
However, sometimes clients ask for more.....so, GRG Health sends out parts of a recorded interview that has been cleaned to prevent identification of the end client or related material information of commercial interest.
Write to us at [email protected] Learn how GRG Health is helping clients gather more in-depth market-level information on such topics
One such example is as follows:
Client Q. So, what kind of work have you done in Oncology?
GRG Health has significant experience in Oncology and can share a presentation to offer more information.
Client Q. Okay great.....but what makes GRG different versus (name's some other company)....?
Well, our team understands the space quite well....and that is clerly reflected in our interviews even.
Client Q. I see.....can you share a sample, please?
Yes, we can share that as an audio file.....
The sample (excerpt from a past in-depth interview conducted by a GRG team member with a key respondent, usually an experienced oncologist):
GRG Interviewer: "Dr. , thank you for your time....we wanted to focus our discussion on gathering your views about p53. I am sure that you already know that p53 is one of the tumour suppressor genes that has been subjected to very comprehensive research and investigation, resulting in some experts referring to it as the guardian of the genome because of its pre-emptive correction of spindle-specific mutations to prevent nondisjunctions!
We wanted to understand your perspective on how this gene is affected by missense mutations.
<Respondent input masked>
GRG Interviewer: "Okay great, Dr. Now, based on your inputs, do you think it is possible that the site of this missense mutation could act as an oncogene and a predictor of therapeutic response?"
<Respondent input masked>
GRG Interviewer: "Okay, Dr. , further to your inputs, do you think an analysis of whole-exome sequencing (WES) output for p53 can yield credible answers? Also what do you think about adding a review of other mutations (including specific driver gene mutations) to that?
<Respondent input masked>
GRG Interviewer: "Okay, Dr. , so if we were to think on the lines of exonal mutations - the coding parts, of course for entities like CDKN2A, FBXW7, NOTCH1, using NGS, what can we expect, according to you?
<Respondent input masked>
GRG Interviewer: "Okay, Dr. , so would we find any in the typical sites like the hotspot in the DNA-binding domain?
<Respondent input masked>
GRG Interviewer: "Okay, Dr. , and what about findings regarding the transactivation domain?
<Respondent input masked>
GRG Interviewer: "Okay, Dr. , so finally, what are your thoughts about the potential correlation between the earlier-discussed and clinical outcomes?
<Respondent input masked>
Usually, the recording is supported with a transcript for the sample to be understood better......
Usually, the recording assures the client about GRG Health as an unusually rigorous research partner.....
Usually, if you leave us a comment (or send us an email), someone from the team reverts within 24 hours to discuss your needs!
0 notes
Text
The role of p53 in anti-tumor immunity and response to immunotherapy
p53 is a transcription factor that regulates the expression of genes involved in tumor suppression. p53 mutations mediate tumorigenesis and occur in approximately 50% of human cancers. p53 regulates hundreds of target genes that induce various cell fates including apoptosis, cell cycle arrest, and DNA damage repair. p53 also plays an important role in anti-tumor immunity by regulating TRAIL, DR5, TLRs, Fas, PKR, ULBP1/2, and CCL2; T-cell inhibitory ligand PD-L1; pro-inflammatory cytokines;... http://dlvr.it/Sty0BW
0 notes
Text
Cancer clinic
Sino Vedic Cancer Clinic - Fight Cancer With Herbs
Sino Vedic Cancer Research Centre was founded in 1985 with the aim to explore the role of herbs to fight cancer. With the experience of over 36 Years and having treated over Fifty Thousand patients, Sino Vedic stands as a pioneer in the field of Herbal Treatment for Cancer. As cancer is a malady of genes & pathways, so the fight against cancer was focused on normalising the cancer cells by reversing their acquired mutations in genes & pathways. To achieve this Sino Vedic Cancer Research Centre has formulated various AYUSH approved Sino Vedic Formulations which help fight cancer without side effects. Cancer clinic
About Cancer - Alternative cancer treatment
A hundred years ago, cancer was not so common, but its incidence has been rising alarmingly since the last couple of decades, probably due to our changing lifestyle and habits. The situation is so alarming that every fourth person has a lifetime risk of cancer. More than 11 lakh new cases of cancer are registered every year in India, whereas this figure is above 14 million worldwide. Alternative treatment for cancer
Cancer hospital - We are constantly exposed to a variety of cancer-causing agents, known as carcinogens, in the food we eat, in the water we drink, and in the air we breathe. Our single meal may contain a dozen of carcinogens in the form of residues of pesticides and insecticides. Exposure to nuclear radiation, ionising radiation (X-rays, Gamma rays), particle radiation emitted by radioactive substances, Solar radiation, and electromagnetic radiation can cause cancer. Likewise, there is a long list of chemical, physical, biological, and geographical carcinogens.
Ayurvedic treatment for cancer - Cancer treatment
The transformation of a normal cell into a cancerous cell is probably not such a critical event in the genesis of cancer; rather it is the inability of immune cells of the body to identify & destroy the newly formed cancer cells when they are few in numbers. We have observed that the risk of cancer is multiplied in those persons, whose immune system is suppressed due to any factor, including chronic stress, old age, chronic debilitating disease, and abuse of drugs such as analgesics, antibiotics, and corticosteroids. Moreover, life has become fast and competitive, from ‘cradle to grave’ leading to chronic stress that further enhances the risk of cancer by suppressing the immune system of the body. The incidence of cancer is higher in persons affected by Human Papilloma Virus (H.P.V.), H.I.V., and other viral infections including Hepatitis B and Hepatitis C.
Cancer is an abnormal growth of cells in any tissue or organ of the body and these cells have a tendency to spread and grow in other parts of the body. Ayurvedic cancer treatment
Normal cell division is a highly regulated mechanism controlled by genes through growth regulatory pathways. Prolonged exposure to carcinogens damages the DNA and induces mutations in the growth regulatory genes including oncogenes (ras, N-myc, c-myc, HER-2/neu, etc), tumour suppressor genes (p53, Rb, Ret, WT-1, APC, etc) and pathways (ras, Rb, myc, etc) leading to loss of control over normal cell division. The mutated cells go haywire and proliferate indiscriminately, usually forming a mass, known as a neoplasm or a malignant tumour or in simple words, a Cancer. Best cancer hospital
Best cancer clinic As time passes, cancer cells go on accumulating further mutations and acquire more evil characteristics such as the ability to invade & move into the adjoining tissues, travel through lymph and blood vessels, lodge and grow in other parts of the body to form colonies (metastasis), create their own blood vessels (tumour angiogenesis) for their nutrition, evade the process of programmed cell death (apoptosis) and acquire the ability of limitless replication, making the cancer cells immortal. By the time most of the cancers are finally diagnosed, they have already added many mutations; for example, ALL (a blood cancer) has been found to have 5 to 10 mutations at the time of diagnosis. Pancreatic cancer has shown 50 to 60 mutations, while Breast & Colon cancers have 50 to 80 mutations at the time of diagnosis. Similarly, most of the cancers have 11 to 15 aberrant (mutated) pathways at the time of diagnosis.
Sino Vedic Formulations - Cancer treatment in india
Treatment for breast cancer - Sino Vedic Cancer Research Centre was founded in 1985 by Dr. S.P. Kaushal, M.B.B.S., M.D. (Chinese Medicine), Ph.D., D.Sc. (Honoris Causa), along with a team of doctors and scientists. Sino Vedic Cancer Research team has identified anticancer herbs having D.N.A. repairing, antimutagenic, antitumourangiogenic, proapoptotic, immunomodulatory, radioprotective & chemoprotective properties. After identification, this team isolated the specific active principles (phytoconstituents) from selected anticancer herbs, which were used to develop various Sino Vedic formulations to fight cancer, without side effects. Treatment for lung cancer
Treatment for prostate cancer Sino Vedic Formulations are better remedies, especially for those patients who have become resistant or refractory to chemotherapy and for those who are not fit to receive chemotherapy and radiotherapy due to old age, marked weakness or any other factor such as kidney and liver dysfunction or cardiac insufficiency. Sino Vedic Formulations help to fight cancer at every step, i.e., genesis, growth and spread of cancer. These formulations tame aggressive cancer cells by repairing damaged D.N.A., inhibiting mutations in genes and pathways, blocking various cancer-promoting enzymes and hormones, reviving the process of apoptosis, inhibiting tumour angiogenesis, and boosting the immune system of the body. Treatment for blood cancer
Sino Vedic Cancer Research Centre has designed this website with the aim to educate people about causes, prevention and early detection of cancer. This website further elaborates the role of Sino Vedic formulations in our fight against Cancer. Treatment for brain tumour
More Information - https://www.sinovedic.com/
0 notes
Text
Mechanism of EGCG binding to p53 revealed: Polyphenolic compounds in green tea may enhance anti-tumor activity by stabilizing p53.
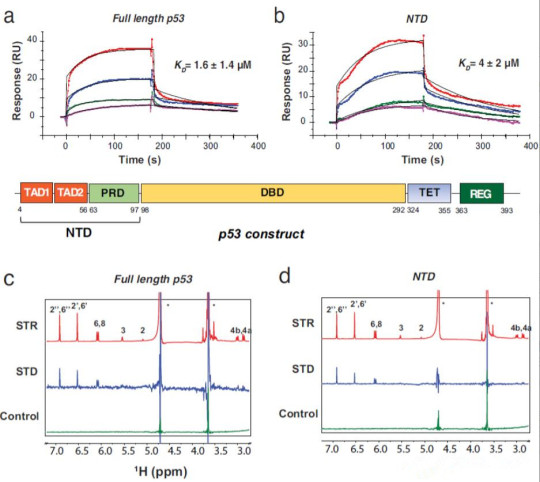
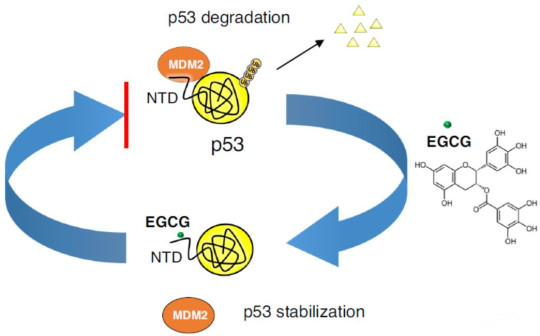
A new study has found that epigallocatechin gallate (EGCG), a polyphenolic compound found in green tea, can induce apoptosis in cancer cells, but its potential molecular mechanisms are not well understood. Recently, a team led by Chunyu Wang at Rensselaer Polytechnic Institute (with China Agricultural University as the first author's affiliation and Jing Zhao as the first author of the paper) published a study in Nature Communications titled "EGCG binds intrinsically disordered N-terminal domain of p53 and disrupts p53-MDM2 interaction". The study used SPR and NMR to report the direct interaction between EGCG and the tumor suppressor p53 (KD = 1.6 ± 1.4 μM), with the disordered N-terminal domain (NTD) identified as the main binding site (KD = 4 ± 2 μM). Large-scale atomic simulations (>100 μs), SAXS, and AUC showed that the EGCG-NTD interaction is dynamic, with EGCG leading to a subpopulation of compact bound conformations. The EGCG-p53 interaction disrupted the interaction between p53 and its regulatory E3 ligase MDM2 and inhibited MDM2-mediated ubiquitination of p53 in vitro, potentially stabilizing p53's anti-tumor activity. Overall, this study provides insights into the anti-cancer mechanism of EGCG and identifies the p53 NTD as a target for cancer drug discovery through dynamic interactions with small molecules.
In recent years, diet-based cancer prevention and treatment have received considerable attention. Green tea is a popular beverage worldwide and has been reported to have inhibitory effects on various types of cancers, such as breast cancer, lung cancer, prostate cancer, and colon cancer. Most of the chemopreventive effects of green tea on cancer are attributed to polyphenolic compounds, with epigallocatechin-3-gallate (EGCG) being the most important. EGCG accounts for 50-80% of the catechins in green tea. A cup of brewed green tea (240 mL) contains 200-300 mg of EGCG. A serum concentration of 0.1-1 μM EGCG can be achieved by drinking a cup of green tea or taking an EGCG tablet. The anticancer effects of EGCG have been confirmed in epidemiological, cell culture and animal studies, as well as clinical trials.
A 10-year prospective study by Nakachi and Imai reported that people who drink more than 10 cups of green tea per day have a lower risk of cancer compared to those who drink less than 3 cups per day. Recently, Shin et al. found in a randomized clinical trial in Korea that green tea extract can reduce the recurrence rate of colorectal adenomas by 44.2%. In vitro, EGCG has been shown to promote growth arrest and induce apoptosis in a variety of human cancer cell lines, including prostate cancer cells, epidermoid carcinoma cells, bladder cancer cells, and colon cancer cells.
In mice, oral administration of green tea or intravenous injection of purified EGCG can inhibit angiogenesis and suppress the growth of solid tumors. At the molecular level, EGCG has been shown to interact with cancer-associated proteins, such as glucose-regulated protein 78 (GRP78) and Ras-GTPase-activating protein SH3 domain-binding protein 1 (G3BP1), with an affinity of approximately μM.
In EGCG-induced apoptosis and growth arrest, p53 plays an important role. p53, often referred to as the "guardian of the genome," is a critical tumor suppressor that is mutated in over 50% of human cancers. p53 promotes cell cycle arrest or apoptosis as a response to cellular stress stimuli such as oxidative stress, oncogene activation, and DNA damage. As a transcription factor, p53 is tightly regulated and has a short half-life. p53 protein is typically maintained at low levels in healthy mammalian cells through continuous ubiquitination and subsequent degradation.
Under cellular stress, ubiquitination of p53 is inhibited, and p53 is stabilized. Then p53 accumulates in the cell nucleus and activates the expression of target genes, thereby triggering cell cycle arrest, apoptosis, and DNA repair processes. In addition to serving as a transcription factor, p53 can also be translocated to the cytoplasm or mitochondria. p53 directly interacts with anti-apoptotic proteins such as Bax and Bcl2 to induce apoptosis and also participates in the anti-aging effects of EGCG.
Full-length p53 consists of an N-terminal domain (NTD), a DNA-binding domain (DBD), a tetramerization domain (TET), and a C-terminal regulatory domain (REG). The NTD is further divided into two transcription activation domains (TAD1 and TAD2) and a proline-rich domain (PRD). The NTD is an intrinsically disordered protein (IDP) that interacts with many proteins and serves as a hub for cellular signal transduction. The NTD is not only necessary for transactivation but also interacts with MDM2 to mediate the ubiquitination and degradation of p53.
Independent of ubiquitination, MDM2 also inhibits transcription by preventing the general transcription factors from binding to the NTD. The apoptotic effect of EGCG on human cancer cells is related to its interference with MDM2-mediated ubiquitination of p53. It has been reported that EGCG can also stabilize p53 and increase the phosphorylation of critical serine residues. In a recent study, EGCG was identified as an inhibitor of the p53-MDM2 interaction from a library of 2,295 plant chemicals. However, the molecular mechanism by which EGCG disrupts the MDM2-p53 interaction is not yet clear.
In this work, direct binding between EGCG and p53 mediated by the NTD of p53 was demonstrated. The study suggests that the EGCG-p53 interaction disrupts the MDM2-p53 interaction and inhibits the ubiquitination of p53, potentially stabilizing p53's anti-tumor activity and providing a structural mechanism for the anti-cancer effects of EGCG.
#EGCG#tea#green tea#cancer cells#apoptosis#molecular mechanisms#p53#N-terminal domain#tumor suppressor#MDM2#ubiquitination#anti-tumor activity#polyphenolic compounds#chemopreventive effects#epidemiological study#cell culture#animal studies#clinical trials#colorectal adenomas#growth arrest、#angiogenesis#solid tumors#cancer-associated proteins#glucose-regulated protein 78 (GRP78)#Ras-GTPase-activating protein SH3 domain-binding protein 1 (G3BP1)#affinity#growth arrest#cellular stress#DNA damage#transcription factor
1 note
·
View note
Text
Drugs that Target p53-Mdm2 Interaction

Drugs that Target p53-Mdm2 Interaction in Biomedical Journal of Scientific & Technical Research
https://biomedres.us/fulltexts/BJSTR.MS.ID.006025.php
The first identified tumor suppressor, p53, is the guardian of the genome and is tightly controlled by its master negative regulator, mouse double minute (Mdm2), and its paralogues Mdm4 and HDMX [1]. p53 is regulated by a negative feedback loop whereby p53 transcriptionally activates Mdm2. In turn, Mdm2 catalyzes the ubiquitination and proteasomal degradation of p53. Virtually all cancers have dysfunctional p53 due to either the gene’s mutation or to dysregulated p53 signaling in the background of wild-type p53. The most common mechanism by which p53 signaling is dysregulated is the overexpression of Mdm2. Its overexpression occurs in tumors either due to gene amplification or by increased transcription and or translation. However, it is unusual for Mdm2 overexpression to be combined with mutated p53 in the same tumor. Consequently, targeting the p53-Mdm2 interaction has become a desirable approach to developing small molecule anticancer drugs.
For more articles in Journals on Biomedical Sciences click here bjstr
Follow on Twitter : https://twitter.com/Biomedres01
Follow on Blogger :https://biomedres01.blogspot.com/
Like Our Pins On : https://www.pinterest.com/biomedres/
#Behavioral Medicine Journals#physical medicine and rehabilitation#Neurological Disorders#journal of biomedical research and reviews impact factor#biomedical science and research journal
0 notes
Link
0 notes