#multicomparison
Explore tagged Tumblr posts
Text
There's something about characters having a swoopy hairstyle that I love



#dreamworks#httyd 2#hiccup haddock#tdp#callum#lol arcane#lol vi#character design#multicomparison#comparison appreciation#my posts
27 notes
·
View notes
Text
Adding some I think of myself and ones that were mentioned in reblog or comments about the post:























"Nice characters are boring" to YOU. I love characters who no matter what, will always have genuine love for humanity in their heart. Characters who dance and laugh and sing with sincerity. Characters who believe in others, and are willing to extend a helping hand to people when no one gave them the same luxury. Characters who have gone through so much but believe, no matter what, that humanity and life is something beautiful and worth protecting
45K notes
·
View notes
Text
MODULE II_Week 1_ ANOVA test_Duration of sleep and happiness
I continue the course with my topic: 'Could the quality of sleep impact happiness?
From the AddHealth code, I focus on variables [H1GH51]: ‘How many hours do you sleep ?’ and [H1FS11]: ‘Do you feel happy ?’
Before continuing with the analysis of variance, I first went back to the fundamentals and managed my variables : Setting variables as numeric, studying the frequency distribution and managing the ‘Refused’ or ‘Don’t know’ response as NAN for both variables.
Code:


I reduce the duration of sleep from 1 to 10 hours for [H1GH51] and check the result by an univariate representation :



[H1FS11]‘Were you happy ? with 0=never or rarely; 1=sometimes; 2=a lot of the time; 3=most of the time or all of the time; 6=Refused; 8=Dont know')
Now that the data are cleaned and managed, I can proceed with analysis of variance. The response variable [H1GH51] is quantitative and the explanatory variable [H1FS11] is categorical with several level of response.
The hypothesis Ho : There is NO relationship between the happiness and duration of sleep and all means are equal ; The alternative H1 is then, There IS a relationship between the happiness and duration of sleep.
I run the ANOVA tests on Python and ask for mean and standard deviation for [H1GH51] (hours of sleep)

And the results are :



The p-Value is 6.96E-20 << 0.05 This would allow us to reject the null hypothesis and say that the duration of sleep and happiness could be linked. But as the explanatory variable [H1FS11] has more than two levels, before concluding, I need to conduct a post hoc test, the TukeyHSD for multicomparison of the means.

And results:

This is very interesting: The ANOVA test revealed a very low p-Value, meaning that duration of sleep and happiness are significantly associated. The postHoc comparisons revealed that in fact, only subjects who responded 0-Never or rarely and 1-sometimes to the question ‘Did you feel happy’ have a different mean . The others means are statistically similar. While this, of course remains statistical, one piece of advice is that if you want to feel happy, sleep more! 😊
0 notes
Text
ANOVA
DataSet: Salary Data based on Experience,Age,Gender,Job Title and Education Level
Issue of the problem:
HO: There is diference in salaly between genders HA: There is a difference is salary between genders
CODE
#Libaries
import numpy as np import pandas as pd import seaborn as sns import matplotlib.pyplot as plt
import scipy.stats as stats import statsmodels.api as sm import statsmodels.formula.api as smf import statsmodels.stats.multicomp as pairwise import statsmodels.stats.multicomp as multicomparison
#main code
data = pd.read_excel('Salary_Data.xlsx') print(data)
boxplt = sns.boxplot(x='Gender', y='Salary', data=data, color=('magenta')).set_title("Gender vs Salary")
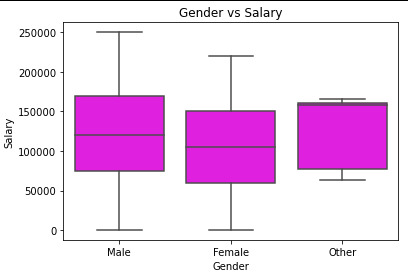
mediangender = data.groupby('Gender').median() print(mediangender)

meangender = data.groupby('Gender').mean() print(meangender)

model = smf.ols(formula='Salary ~ C(Gender)',data=data) res = model.fit() print(res.summary())
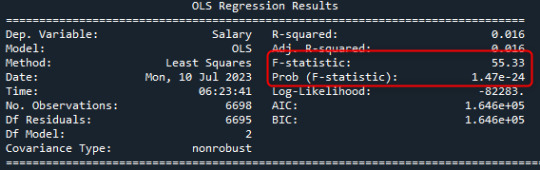
After verifying the p-value < 0.05 with ANOVA, it is necessery to verify which group is different. ANOVA can not give this information. For that we performe a POST HOC TEST which conducts a post hoc comparisions. From serveral choices, for thsi dataset, I sused the tukey. Tukey -> conduct a tukey honest significant difference test mc=multicomparison.MultiComparison((data['Salary']),data['Gender']) result = mc.tukeyhsd(0.05) print(result.summary())

By analysing the data:
->P-value for the difference in means between Female and Male: 0.001
->P-value for the difference in means between Female and Other: 0.4072
->P-value for the difference in means between Male and Other: 0.9
Thus, we would conclude that there is a statistically significant difference between the means of Female and Male, but not a statistically significant difference between the means of Female and Other, Male and Others.
1 note
·
View note
Text
Lovable Fictional Blue Colored Amphibious Characters



#dc#dcu#nina mazursky#gdt hellboy#abe sapien#the shape of water#colors#multicomparison#comparison appreciation#my posts#tsow amphibian man
43 notes
·
View notes
Text
Mama Odie from Disney's The Princess and the Frog

Luz Noceda from Disney's The Owl House

Prudence Night from the Chilling Adventures of Sabrina

Black Witches










Black Witches
Tabby || The Craft: Legacy
Nynaeve al'Meara || Wisdom || The Wheel of Time
Calypso || Sea Goddess/Witch || Pirates of The Carribean
Jada Sheilds || Whitelighter-Witch || Charmed (2018)
Leta Lestrange || Fantastic Beasts: Crimes of Grindelwald
Eulalie Hicks || Fantastic Beasts: Secrets of Dumbledore
Fringilla Vigo || The Witcher
Seraphina Picquery || Fantastic Beasts and Where to Find Them
Kaela Danso || Charmed (2018)
Macy Vaughn and Maggie Vera || Charmed (2018)
873 notes
·
View notes
Text
Assignment 1
Assignment 1
This assignment is aim to test differences in the mean number of cigarettes smoked among young adults of 5 different levels of health condition. The data for this study is from U.S. National Epidemiological Survey on Alcohol and Related Conditions (NESARC). The response variable “NUMCIGMO_EST” is the number cigarettes smoked in a month based on the young adults age 18 to 25 who have smoked in the past 12 month. NUMCIGMO_EST is created as follows: 1. “USFREQMO” is converted to number of days smoked in the past month, 2. multiplying new “USFREQMO” and “S3AQ3C1" which is cigarettes/day. The explanatory variable in the test is column “S1Q16”, which is described as “SELF-PERCEIVED CURRENT HEALTH”, with values from 1 (excellent) to 5 (poor). Since this is a C->Q test, thus I ran ANOVA using OLS model to calculate F-statistic and p-value,
Here is the output from the OLS output:
=========================================================================== Dep. Variable: NUMCIGMO_EST R-squared: 0.031 Model: OLS Adj. R-squared: 0.025 Method: Least Squares F-statistic: 5.001 Date: Mon, 22 Mar 2021 Prob (F-statistic): 0.000567 Time: 19:54:15 Log-Likelihood: -4479.1 No. Observations: 629 AIC: 8968. Df Residuals: 624 BIC: 8991. Df Model: 4 Covariance Type: nonrobust =========================================================================== coef std err t P>|t| [0.025 0.975] ----------------------------------------------------------------------------------- Intercept 315.4803 22.538 13.997 0.000 271.220 359.741 C(S1Q16)[T.2.0] 15.4845 30.537 0.507 0.612 -44.483 75.452 C(S1Q16)[T.3.0] 89.5654 31.530 2.841 0.005 27.648 151.482 C(S1Q16)[T.4.0] 119.1863 50.173 2.376 0.018 20.658 217.715 C(S1Q16)[T.5.0] 348.8054 115.867 3.010 0.003 121.268 576.342 =========================================================================== Omnibus: 346.462 Durbin-Watson: 2.001 Prob(Omnibus): 0.000 Jarque-Bera (JB): 4387.366 Skew: 2.167 Prob(JB): 0.00 Kurtosis: 15.191 Cond. No. 10.7 ===========================================================================
With F-statistic is 5.001 and p-value is 0,000567, I can safely reject the null hypothesis and conclude that there is an association between number of cigarettes smoked and the health conditions. To determine which levels of health conditions are different from the others, I perform a post hoc test using the Tukey HSDT, or Honestly Significant Difference Test (this is implemented by calling “MultiComparison” function). The following shows the result of the Post Hoc Pair Comparison:
Multiple Comparison of Means - Tukey HSD, FWER=0.05 ======================================================= group1 group2 meandiff p-adj lower upper reject ------------------------------------------------------- 1.0 2.0 15.4845 0.9 -68.0574 99.0263 False 1.0 3.0 89.5654 0.0374 3.3073 175.8234 True 1.0 4.0 119.1863 0.1236 -18.0761 256.4488 False 1.0 5.0 348.8054 0.0227 31.8177 665.7931 True 2.0 3.0 74.0809 0.1025 -8.4764 156.6383 False 2.0 4.0 103.7019 0.2205 -31.2657 238.6694 False 2.0 5.0 333.3209 0.0328 17.3202 649.3216 True 3.0 4.0 29.621 0.9 -107.0445 166.2864 False 3.0 5.0 259.24 0.1668 -57.4896 575.9697 False 4.0 5.0 229.619 0.3295 -104.6237 563.8618 False -------------------------------------------------------
In the last column, we can determine which health level of groups smoke significantly different mean number of cigarettes than the others by identifying the comparisons in which we can reject the null hypothesis, that is, in which reject equals true.
Python script for the homework:
import numpy import pandas import statsmodels.formula.api as smf import statsmodels.stats.multicomp as multi
data = pandas.read_csv('nesarc.csv', low_memory=False)
data['S3AQ3B1'] = pandas.to_numeric(data['S3AQ3B1'], errors="coerce") data['S3AQ3C1'] = pandas.to_numeric(data['S3AQ3C1'], errors="coerce") data['CHECK321'] = pandas.to_numeric(data['CHECK321'], errors="coerce")
#subset data to young adults age 18 to 25 who have smoked in the past 12 months sub1=data[(data['AGE']>=18) & (data['AGE']<=25) & (data['CHECK321']==1)]
#SETTING MISSING DATA sub1['S3AQ3B1']=sub1['S3AQ3B1'].replace(9, numpy.nan) sub1['S3AQ3C1']=sub1['S3AQ3C1'].replace(99, numpy.nan)
#recoding number of days smoked in the past month recode1 = {1: 30, 2: 22, 3: 14, 4: 5, 5: 2.5, 6: 1} sub1['USFREQMO']= sub1['S3AQ3B1'].map(recode1)
#converting new variable USFREQMMO to numeric sub1['USFREQMO']= pandas.to_numeric(sub1['USFREQMO'], errors="coerce")
sub1['NUMCIGMO_EST']=sub1['USFREQMO'] * sub1['S3AQ3C1']
sub1['NUMCIGMO_EST']= pandas.to_numeric(sub1['NUMCIGMO_EST'], errors="coerce")
sub1['S1Q16']=sub1['S1Q16'].replace(9, numpy.nan) sub3 = sub1[['NUMCIGMO_EST', 'S1Q16']].dropna()
''' By running an analysis of variance, we're asking whether the number of cigarettes smoked differs for different health conditions. ''' model2 = smf.ols(formula='NUMCIGMO_EST ~ C(S1Q16)', data=sub3).fit() print (model2.summary())
print ('means for numcigmo_est by major depression status') m2= sub3.groupby('S1Q16').mean() print (m2)
print ('standard deviations for numcigmo_est by major depression status') sd2 = sub3.groupby('S1Q16').std() print (sd2)
mc1 = multi.MultiComparison(sub3['NUMCIGMO_EST'], sub3['S1Q16']) res1 = mc1.tukeyhsd() print(res1.summary())
1 note
1 note
·
View note
Text
Week 1 Assignment – Running an Analysis of variance (ANOVA)
Objective:
The assignment of the week deals with Analysis of variance. Given a dataset some form of Statistical Analysis test to be performed to check and evaluate its Statistical significance.
Before getting into the crux of the problem let us understand some of the important concepts
Hypothesis testing - It is one of the most important Inferential Statistics where the hypothesis is performed on the sample data from a larger population. Hypothesis testing is a statistical assumption taken by an analyst on the nature of the data and its reason for analysis. In other words, Statistical Hypothesis testing assesses evidence provided by data in favour of or against each hypothesis about the problem.
There are two types of Hypothesis
Null Hypothesis – The null hypothesis is assumed to be true until evidence indicate otherwise. The general assumptions made on the data (people with depression are more likely to smoke)
Alternate Hypothesis – Once stronger evidences are made, one can reject Null hypothesis and accept Alternate hypothesis (One needs to come with strong evidence to challenge the null hypothesis and draw proper conclusions).In this case one needs to show evidences such that there is no relation between people smoking and their depression levels
Example:
The Null hypothesis is that the number of cigarettes smoked by the person is dependent on the person’s depression level. Based on the p-value we make conclusions either to accept the null hypothesis or fail to accept the null hypothesis(accept alternative hypothesis)
Steps involved in Hypothesis testing:
1. Choose the Null hypothesis (H0 ) and alternate hypothesis (Ha)
2. Choose the sample
3. Assess the evidence
4. Draw the conclusions
The Null hypothesis is accepted/rejected based on the p-value significance level of test
If p<= 0.05, then reject the null hypothesis (accept the alternate hypothesis)
If p > 0.05 null hypothesis is accepted
Wrongly rejecting the null hypothesis leads to type one error
Sampling variability:
The measures of the sample (subset of population) varying from the measures of the population is called Sampling variability. In other words sample results changing from sample to sample.
Central Limit theorem:
As long as adequately large samples and an adequately large number of samples are used from a population,the distribution of the statistics of the sample will be normally distributed. In other words, the more the sample the accurate it is to the population parameters.
Choosing Statistical test:
Please find the below tabulation to identify what test can be done at a given condition. Some of the statistical tools used for this are
Chi-square test of independence, ANOVA- Analysis of variance, correlation coefficient.
Explanatory variables are input or independent variable And response variable is the output variable or dependent variable.
Explanatory Response Type of test
Categorical Categorical Chi-square test
Quantitative Quantitative Pearson correlation
Categorical Quantitative ANOVA
Quantitative Categorical Chi-square test
ANOVA:
Anova F test, helps to identify, Are the difference among the sample means due to true difference among the population or merely due to sampling variability.
F = variation among sample means / by variations within groups
Let’s implement our learning in python.
The Null hypothesis here is smoking and depression levels are unrelated
The Alternate hypothesis is smoking and depression levels are related.
# importing required libraries
import numpy as np
import pandas as pd
import statsmodels.formula.api as smf
data = pd.read_csv("my_data...nesarc.csv",low_memory=False)
#setting variables you will be working with to numeric
data['S3AQ3B1'] = data['S3AQ3B1'].convert_objects(convert_numeric=True)
#data['S3AQ3B1'] = pd.to_numeric(data.S3AQ3B1)
data['S3AQ3C1'] = data['S3AQ3C1'].convert_objects(convert_numeric=True)
#data['S3AQ3C1'] = pd.to_numeric(data.S3AQ3C1)
data['CHECK321'] = data['CHECK321'].convert_objects(convert_numeric=True)
#data['CHECK321'] = pd.to_numeric(data.CHECK321)
#subset data to young adults age 18 to 25 who have smoked in the past 12 months
sub1=data[(data['AGE']>=18) & (data['AGE']<=25) & (data['CHECK321']==1)]
#SETTING MISSING DATA
sub1['S3AQ3B1']=sub1['S3AQ3B1'].replace(9, np.nan)
sub1['S3AQ3C1']=sub1['S3AQ3C1'].replace(99, np.nan)
#recoding number of days smoked in the past month
recode1 = {1: 30, 2: 22, 3: 14, 4: 5, 5: 2.5, 6: 1}
sub1['USFREQMO']= sub1['S3AQ3B1'].map(recode1)
#converting new variable USFREQMMO to numeric
sub1['USFREQMO']= sub1['USFREQMO'].convert_objects(convert_numeric=True)
# Creating a secondary variable multiplying the days smoked/month and the number of cig/per day
sub1['NUMCIGMO_EST']=sub1['USFREQMO'] * sub1['S3AQ3C1']
sub1['NUMCIGMO_EST']= sub1['NUMCIGMO_EST'].convert_objects(convert_numeric=True)
ct1 = sub1.groupby('NUMCIGMO_EST').size()
print (ct1)
print(sub1['MAJORDEPLIFE'])
# using ols function for calculating the F-statistic and associated p value
The ols is the ordinary least square function takes the response variable NUMCIGMO_EST and its explanatory variable MAJORDEPLIFE, C is indicated to specify that it’s a categorical variable.
model1 = smf.ols(formula='NUMCIGMO_EST ~ C(MAJORDEPLIFE)', data=sub1)
results1 = model1.fit()
print (results1.summary())
Inference – Since the p value is greater than 0.05, we accept the null hypothesis that the sample means are statistically equal. And there is no association between depression levels and cigarette smoked.
sub2 = sub1[['NUMCIGMO_EST', 'MAJORDEPLIFE']].dropna()
print ('means for numcigmo_est by major depression status')
m1= sub2.groupby('MAJORDEPLIFE').mean()
print (m1)
print ('standard deviations for numcigmo_est by major depression status')
sd1 = sub2.groupby('MAJORDEPLIFE').std()
print (sd1)
Since the null hypothesis is true we conclude that the sample mean and standard deviation of the two samples are statistically equal. From the sample statistics we infer that the depression levels and smoking are unrelated to each other
Till now, we ran the ANOVA test for two levels of the categorical variable(MAJORDEPLIFE, 0,1) Lets now look at the categorical variable(ETHRACE2A,White, Black, American Indian, Asian native and Latino) having five levels and see how the differences in the mean are captured.
The problem here is, The F-statistic and ‘p’ value does not provide any insight as to why the null hypothesis is rejected when there are multiple levels in categorical Explanatory variable.
The significant ANOVA does not tell which groups are different from the others. To determine this we would need to perform post-hoc test for ANOVA
Why Post-hoc tests for ANOVA?
Post hoc test known as after analysis test, This is performed to prevent excessive Type one error. Not implementing this leads to family wise error rate, given by the formula
FWE = 1 – (1 – αIT)C
Where:
αIT = alpha level for an individual test (e.g. .05),
c = Number of comparisons.
Post-hoc tests are designed to evaluate the difference between the pair of means while protecting against inflation of type one error.
Let’s continue the code to perform post-hoc test.
# Importing the library
Import statsmodels.stats.multicomp as multi
# adding the variable of interest to a separate data frame
sub3 = sub1[['NUMCIGMO_EST', 'ETHRACE2A']].dropna()
# calling the ols function and passing the explanatory categorical and response variable
model2 = smf.ols(formula='NUMCIGMO_EST ~ C(ETHRACE2A)', data=sub3).fit()
print(model2.summary())
print ('means for numcigmo_est by major depression status')
m2= sub3.groupby('ETHRACE2A').mean()
print(m2)
print('standard deviations for numcigmo_est by major depression status')
sd2 = sub3.groupby('ETHRACE2A').std()
print(sd2)
# Include required parameters in the MultiComparison function and then run the post-hoc TukeyHSD test
mc1 = multi.MultiComparison(sub3['NUMCIGMO_EST'], sub3['ETHRACE2A'])
res1 = mc1.tukeyhsd()
print(res1.summary())
1 note
·
View note
Photo
2022 tiger tigers year of the tiger aladdin teenage mutant ninja turtles transformers the jungle book thundercats arlo the alligator boy kung fu panda marvel comics winnie the pooh beast wars tmnt 2012 rajah aladdin rajah tiger claw tmnt tiger claw tigatron shere khan tygra alia arlo alia master tigress tigress tigra greer grant tigger









Year of the Tiger
#celebrations#tigers#multicomparison#disney#aladdin 1992#tmnt 2012#transformers#the jungle book 1967#thundercats#dreamworks#kfp#marvel#winnie the pooh
479 notes
·
View notes
Text
Running an ANOVA
Having selected the GapMinder data set and a research question, managed my variables of interest, namely oil consumption per person, residential electricity consumption per person and urban population, and visualized their relationship graphically, we are now ready to test these relationships statistically.
The analysis of variance (ANOVA) assesses whether the means of two or more groups of categorical variables are statistically different from each other. Since all of the variables chosen are quantitative, they are categorised for the purpose of running the test. Post hoc paired comparisons were also carried out in instances where the original statistical test was significant for explanatory variables with more than two levels of categories.
The codes written for this program are shown below:
#############################################
# Import required libraries import pandas as pd import numpy as np import seaborn as sns import matplotlib as mpl import matplotlib.pyplot as plt import statsmodels.formula.api as smf import statsmodels.stats.multicomp as multi
# Bug fix for display formats and change settings to show all rows and columns pd.set_option('display.float_format', lambda x:'%f'%x) pd.set_option('display.max_columns', None) pd.set_option('display.max_rows', None)
# Read in the GapMinder dataset raw_data = pd.read_csv('./gapminder.csv', low_memory=False)
# Report facts regarding the original dataset print("Facts regarding the original GapMinder dataset:") print("---------------------------------------") print("Number of countries: {0}".format(len(raw_data))) print("Number of variables: {0}\n".format(len(raw_data.columns))) print("All variables:\n{0}\n".format(list(raw_data.columns))) print("Data types of each variable:\n{0}\n".format(raw_data.dtypes)) print("First 5 rows of entries:\n{0}\n".format(raw_data.head())) print("=====================================\n")
# Choose variables of interest # var_of_int = ['country', 'incomeperperson', 'alcconsumption', 'co2emissions', # 'internetuserate', 'oilperperson', 'relectricperperson', 'urbanrate'] var_of_int = ['oilperperson', 'relectricperperson', 'urbanrate'] print("Chosen variables of interest:\n{0}\n".format(var_of_int)) print("=====================================\n")
# Code out missing values by replacing with NumPy's NaN data type data = raw_data[var_of_int].replace(' ', np.nan) print("Replaced missing values with NaNs:\n{0}\n".format(data.head())) print("=====================================\n")
# Cast the numeric variables to the appropriate data type then quartile split numeric_vars = var_of_int[:] for var in numeric_vars: data[var] = pd.to_numeric(data[var], downcast='float', errors='raise') print("Simple statistics of each variable:\n{0}\n".format(data.describe())) print("=====================================\n")
# Create secondary variables to investigate frequency distributions print("Separate continuous values categorically using secondary variables:") print("---------------------------------------") data['oilpp (tonnes)'] = pd.cut(data['oilperperson'], 4) oil_val_count = data.groupby('oilpp (tonnes)').size() oil_dist = data['oilpp (tonnes)'].value_counts(sort=False, dropna=True, normalize=True) oil_freq_tab = pd.concat([oil_val_count, oil_dist], axis=1) oil_freq_tab.columns = ['value_count', 'frequency'] print("Frequency table of oil consumption per person:\n{0}\n".format(oil_freq_tab))
data['relectricpp (kWh)'] = pd.cut(data['relectricperperson'], 4) # Variable range is extended by 0.1% elec_val_count = data.groupby('relectricpp (kWh)').size() elec_dist = data['relectricpp (kWh)'].value_counts(sort=False, dropna=True, normalize=True) elec_freq_tab = pd.concat([elec_val_count, elec_dist], axis=1) elec_freq_tab.columns = ['value_count', 'frequency'] print("Frequency table of residential electricity consumption per person:\n{0}\n".format(elec_freq_tab))
data['urbanr (%)'] = pd.cut(data['urbanrate'], 4) urb_val_count = data.groupby('urbanr (%)').size() urb_dist = data['urbanr (%)'].value_counts(sort=False, dropna=True, normalize=True) urb_freq_tab = pd.concat([urb_val_count, urb_dist], axis=1) urb_freq_tab.columns = ['value_count', 'frequency'] print("Frequency table of urban population:\n{0}\n".format(urb_freq_tab)) print("=====================================\n")
# Code in valid data in place of missing data for each variable print("Number of missing data in variables:") print("oilperperson: {0}".format(data['oilperperson'].isnull().sum())) print("relectricperperson: {0}".format(data['relectricperperson'].isnull().sum())) print("urbanrate: {0}\n".format(data['urbanrate'].isnull().sum())) print("=====================================\n")
print("Investigate entries with missing urbanrate data:\n{0}\n".format(data[['oilperperson', 'relectricperperson']][data['urbanrate'].isnull()])) print("Data for other variables are also missing for 90% of these entries.") print("Therefore, eliminate them from the dataset.\n") data = data[data['urbanrate'].notnull()] print("=====================================\n")
null_elec_data = data[data['relectricperperson'].isnull()].copy() print("Investigate entries with missing relectricperperson data:\n{0}\n".format(null_elec_data.head())) elec_map_dict = data.groupby('urbanr (%)').median()['relectricperperson'].to_dict() print("Median values of relectricperperson corresponding to each urbanrate group:\n{0}\n".format(elec_map_dict)) null_elec_data['relectricperperson'] = null_elec_data['urbanr (%)'].map(elec_map_dict) data = data.combine_first(null_elec_data) data['relectricpp (kWh)'] = pd.cut(data['relectricperperson'], 4) print("Replace relectricperperson NaNs based on their quartile group's median:\n{0}\n".format(data.head())) print("-------------------------------------\n")
null_oil_data = data[data['oilperperson'].isnull()].copy() oil_map_dict = data.groupby('urbanr (%)').median()['oilperperson'].to_dict() print("Median values of oilperperson corresponding to each urbanrate group:\n{0}\n".format(oil_map_dict)) null_oil_data['oilperperson'] = null_oil_data['urbanr (%)'].map(oil_map_dict) data = data.combine_first(null_oil_data) data['oilpp (tonnes)'] = pd.cut(data['oilperperson'], 4) print("Replace oilperperson NaNs based on their quartile group's median:\n{0}\n".format(data.head())) print("=====================================\n")
# Investigate the new frequency distributions print("Report the new frequency table for each variable:") print("---------------------------------------") oil_val_count = data.groupby('oilpp (tonnes)').size() oil_dist = data['oilpp (tonnes)'].value_counts(sort=False, dropna=True, normalize=True) oil_freq_tab = pd.concat([oil_val_count, oil_dist], axis=1) oil_freq_tab.columns = ['value_count', 'frequency'] print("Frequency table of oil consumption per person:\n{0}\n".format(oil_freq_tab))
elec_val_count = data.groupby('relectricpp (kWh)').size() elec_dist = data['relectricpp (kWh)'].value_counts(sort=False, dropna=True, normalize=True) elec_freq_tab = pd.concat([elec_val_count, elec_dist], axis=1) elec_freq_tab.columns = ['value_count', 'frequency'] print("Frequency table of residential electricity consumption per person:\n{0}\n".format(elec_freq_tab))
urb_val_count = data.groupby('urbanr (%)').size() urb_dist = data['urbanr (%)'].value_counts(sort=False, dropna=True, normalize=True) urb_freq_tab = pd.concat([urb_val_count, urb_dist], axis=1) urb_freq_tab.columns = ['value_count', 'frequency'] print("Frequency table of urban population:\n{0}\n".format(urb_freq_tab)) print("=====================================\n")
# Run ANOVA print("ANOVA results:") print("-------------------------------------") model1 = smf.ols(formula='relectricperperson ~ C(Q("oilpp (tonnes)"))', data=data) m1, std1 = data[['relectricperperson', 'oilpp (tonnes)']].groupby('oilpp (tonnes)').mean(), data[['relectricperperson', 'oilpp (tonnes)']].groupby('oilpp (tonnes)').std() mc1 = multi.MultiComparison(data['relectricperperson'], data['oilpp (tonnes)']) print("relectricperperson ~ oilpp (tonnes)\n{0}".format(model1.fit().summary())) print("Means for relectricperperson by oilpp status:\n{0}\n".format(m1)) print("Standard deviations for relectricperperson by oilpp status:\n{0}\n".format(std1)) print("MultiComparison summary:\n{0}\n".format(mc1.tukeyhsd().summary()))
model2 = smf.ols(formula='relectricperperson ~ C(Q("urbanr (%)"))', data=data) m2, std2 = data[['relectricperperson', 'urbanr (%)']].groupby('urbanr (%)').mean(), data[['relectricperperson', 'urbanr (%)']].groupby('urbanr (%)').std() mc2 = multi.MultiComparison(data['relectricperperson'], data['urbanr (%)']) print("relectricperperson ~ urbanr (%)\n{0}".format(model2.fit().summary())) print("Means for relectricperperson by urbanr status:\n{0}\n".format(m2)) print("Standard deviations for relectricperperson by urbanr status:\n{0}\n".format(std2)) print("MultiComparison summary:\n{0}\n".format(mc2.tukeyhsd().summary()))
#############################################
The output of the program is as follow:
#############################################
Facts regarding the original GapMinder dataset: --------------------------------------- Number of countries: 213 Number of variables: 16
All variables: ['country', 'incomeperperson', 'alcconsumption', 'armedforcesrate', 'breastcancerper100th', 'co2emissions', 'femaleemployrate', 'hivrate', 'internetuserate', 'lifeexpectancy', 'oilperperson', 'polityscore', 'relectricperperson', 'suicideper100th', 'employrate', 'urbanrate']
Data types of each variable: country object incomeperperson object alcconsumption object armedforcesrate object breastcancerper100th object co2emissions object femaleemployrate object hivrate object internetuserate object lifeexpectancy object oilperperson object polityscore object relectricperperson object suicideper100th object employrate object urbanrate object dtype: object
First 5 rows of entries: country incomeperperson alcconsumption armedforcesrate \ 0 Afghanistan .03 .5696534 1 Albania 1914.99655094922 7.29 1.0247361 2 Algeria 2231.99333515006 .69 2.306817 3 Andorra 21943.3398976022 10.17 4 Angola 1381.00426770244 5.57 1.4613288
breastcancerper100th co2emissions femaleemployrate hivrate \ 0 26.8 75944000 25.6000003814697 1 57.4 223747333.333333 42.0999984741211 2 23.5 2932108666.66667 31.7000007629394 .1 3 4 23.1 248358000 69.4000015258789 2
internetuserate lifeexpectancy oilperperson polityscore \ 0 3.65412162280064 48.673 0 1 44.9899469578783 76.918 9 2 12.5000733055148 73.131 .42009452521537 2 3 81 4 9.99995388324075 51.093 -2
relectricperperson suicideper100th employrate urbanrate 0 6.68438529968262 55.7000007629394 24.04 1 636.341383366604 7.69932985305786 51.4000015258789 46.72 2 590.509814347428 4.8487696647644 50.5 65.22 3 5.36217880249023 88.92 4 172.999227388199 14.5546770095825 75.6999969482422 56.7
=====================================
Chosen variables of interest: ['oilperperson', 'relectricperperson', 'urbanrate']
=====================================
Replaced missing values with NaNs: oilperperson relectricperperson urbanrate 0 NaN NaN 24.04 1 NaN 636.341383366604 46.72 2 .42009452521537 590.509814347428 65.22 3 NaN NaN 88.92 4 NaN 172.999227388199 56.7
=====================================
Simple statistics of each variable: oilperperson relectricperperson urbanrate count 63.000000 136.000000 203.000000 mean 1.484085 1173.179199 56.769348 std 1.825090 1681.440430 23.844936 min 0.032281 0.000000 10.400000 25% 0.532542 203.652103 36.830000 50% 1.032470 597.136444 57.939999 75% 1.622737 1491.145233 74.209999 max 12.228645 11154.754883 100.000000
=====================================
Separate continuous values categorically using secondary variables: --------------------------------------- Frequency table of oil consumption per person: value_count frequency oilpp (tonnes) (0.0201, 3.0814] 58 0.920635 (3.0814, 6.13] 3 0.047619 (6.13, 9.18] 1 0.015873 (9.18, 12.229] 1 0.015873
Frequency table of residential electricity consumption per person: value_count frequency relectricpp (kWh) (-11.155, 2788.689] 122 0.897059 (2788.689, 5577.377] 10 0.073529 (5577.377, 8366.0662] 3 0.022059 (8366.0662, 11154.755] 1 0.007353
Frequency table of urban population: value_count frequency urbanr (%) (10.31, 32.8] 42 0.206897 (32.8, 55.2] 51 0.251232 (55.2, 77.6] 68 0.334975 (77.6, 100] 42 0.206897
=====================================
Number of missing data in variables: oilperperson: 150 relectricperperson: 77 urbanrate: 10
=====================================
Investigate entries with missing urbanrate data: oilperperson relectricperperson 43 nan nan 71 nan 0.000000 75 nan nan 121 nan nan 134 nan nan 143 nan nan 157 nan nan 170 nan nan 187 2.006515 1831.731812 198 nan nan
Data for other variables are also missing for 90% of these entries. Therefore, eliminate them from the dataset.
=====================================
Investigate entries with missing relectricperperson data: oilperperson relectricperperson urbanrate oilpp (tonnes) \ 0 nan nan 24.040001 NaN 3 nan nan 88.919998 NaN 5 nan nan 30.459999 NaN 8 nan nan 46.779999 NaN 12 nan nan 83.699997 NaN
relectricpp (kWh) urbanr (%) 0 NaN (10.31, 32.8] 3 NaN (77.6, 100] 5 NaN (10.31, 32.8] 8 NaN (32.8, 55.2] 12 NaN (77.6, 100]
Median values of relectricperperson corresponding to each urbanrate group: {'(10.31, 32.8]': 59.848274, '(32.8, 55.2]': 278.73962, '(55.2, 77.6]': 753.20978, '(77.6, 100]': 1741.4866}
Replace relectricperperson NaNs based on their quartile group's median: oilperperson relectricperperson urbanrate oilpp (tonnes) \ 0 nan 59.848274 24.040001 NaN 1 nan 636.341370 46.720001 NaN 2 0.420095 590.509827 65.220001 (0.0201, 3.0814] 3 nan 1741.486572 88.919998 NaN 4 nan 172.999222 56.700001 NaN
relectricpp (kWh) urbanr (%) 0 (-11.155, 2788.689] (10.31, 32.8] 1 (-11.155, 2788.689] (32.8, 55.2] 2 (-11.155, 2788.689] (55.2, 77.6] 3 (-11.155, 2788.689] (77.6, 100] 4 (-11.155, 2788.689] (55.2, 77.6]
-------------------------------------
Median values of oilperperson corresponding to each urbanrate group: {'(10.31, 32.8]': 0.079630107, '(32.8, 55.2]': 0.35917261, '(55.2, 77.6]': 0.84457392, '(77.6, 100]': 2.0878479}
Replace oilperperson NaNs based on their quartile group's median: oilperperson relectricperperson urbanrate oilpp (tonnes) \ 0 0.079630 59.848274 24.040001 (0.0201, 3.0814] 1 0.359173 636.341370 46.720001 (0.0201, 3.0814] 2 0.420095 590.509827 65.220001 (0.0201, 3.0814] 3 2.087848 1741.486572 88.919998 (0.0201, 3.0814] 4 0.844574 172.999222 56.700001 (0.0201, 3.0814]
relectricpp (kWh) urbanr (%) 0 (-11.155, 2788.689] (10.31, 32.8] 1 (-11.155, 2788.689] (32.8, 55.2] 2 (-11.155, 2788.689] (55.2, 77.6] 3 (-11.155, 2788.689] (77.6, 100] 4 (-11.155, 2788.689] (55.2, 77.6]
=====================================
Report the new frequency table for each variable: --------------------------------------- Frequency table of oil consumption per person: value_count frequency oilpp (tonnes) (0.0201, 3.0814] 198 0.975369 (3.0814, 6.13] 3 0.014778 (6.13, 9.18] 1 0.004926 (9.18, 12.229] 1 0.004926
Frequency table of residential electricity consumption per person: value_count frequency (-11.155, 2788.689] 189 0.931034 (2788.689, 5577.377] 10 0.049261 (5577.377, 8366.0662] 3 0.014778 (8366.0662, 11154.755] 1 0.004926
Frequency table of urban population: value_count frequency (10.31, 32.8] 42 0.206897 (32.8, 55.2] 51 0.251232 (55.2, 77.6] 68 0.334975 (77.6, 100] 42 0.206897
=====================================
ANOVA results: ------------------------------------- relectricperperson ~ oilpp (tonnes) OLS Regression Results ============================================================================== Dep. Variable: relectricperperson R-squared: 0.350 Model: OLS Adj. R-squared: 0.340 Method: Least Squares F-statistic: 35.72 Date: Mon, 10 Aug 2020 Prob (F-statistic): 1.63e-18 Time: 02:21:51 Log-Likelihood: -1721.6 No. Observations: 203 AIC: 3451. Df Residuals: 199 BIC: 3464. Df Model: 3 Covariance Type: nonrobust ============================================================================================================ coef std err t P>|t| [95.0% Conf. Int.] ------------------------------------------------------------------------------------------------------------ Intercept 830.4105 83.731 9.918 0.000 665.298 995.523 C(Q("oilpp (tonnes)"))[T.(3.0814, 6.13]] 5618.9698 685.364 8.199 0.000 4267.462 6970.477 C(Q("oilpp (tonnes)"))[T.(6.13, 9.18]] 7532.1579 1181.164 6.377 0.000 5202.953 9861.362 C(Q("oilpp (tonnes)"))[T.(9.18, 12.229]] 634.4268 1181.164 0.537 0.592 -1694.778 2963.631 ============================================================================== Omnibus: 141.891 Durbin-Watson: 1.778 Prob(Omnibus): 0.000 Jarque-Bera (JB): 1183.713 Skew: 2.690 Prob(JB): 9.12e-258 Kurtosis: 13.536 Cond. No. 14.3 ==============================================================================
Warnings: [1] Standard Errors assume that the covariance matrix of the errors is correctly specified. Means for relectricperperson by oilpp status: relectricperperson oilpp (tonnes) (0.0201, 3.0814] 830.410461 (3.0814, 6.13] 6449.380371 (6.13, 9.18] 8362.568359 (9.18, 12.229] 1464.837280
Standard deviations for relectricperperson by oilpp status: relectricperperson oilpp (tonnes) (0.0201, 3.0814] 1108.687744 (3.0814, 6.13] 4128.518555 (6.13, 9.18] nan (9.18, 12.229] nan
MultiComparison summary: Multiple Comparison of Means - Tukey HSD,FWER=0.05 ======================================================================= group1 group2 meandiff lower upper reject ----------------------------------------------------------------------- (0.0201, 3.0814] (3.0814, 6.13] 5618.9698 3843.1866 7394.753 True (0.0201, 3.0814] (6.13, 9.18] 7532.1579 4471.7516 10592.5642 True (0.0201, 3.0814] (9.18, 12.229] 634.4268 -2425.9795 3694.8331 False (3.0814, 6.13] (6.13, 9.18] 1913.1881 -1611.7745 5438.1507 False (3.0814, 6.13] (9.18, 12.229] -4984.543 -8509.5056 -1459.5804 True (6.13, 9.18] (9.18, 12.229] -6897.7311 -11214.911 -2580.5512 True -----------------------------------------------------------------------
relectricperperson ~ urbanr (%) OLS Regression Results ============================================================================== Dep. Variable: relectricperperson R-squared: 0.320 Model: OLS Adj. R-squared: 0.310 Method: Least Squares F-statistic: 31.26 Date: Mon, 10 Aug 2020 Prob (F-statistic): 1.33e-16 Time: 02:21:51 Log-Likelihood: -1726.1 No. Observations: 203 AIC: 3460. Df Residuals: 199 BIC: 3473. Df Model: 3 Covariance Type: nonrobust ====================================================================================================== coef std err t P>|t| [95.0% Conf. Int.] ------------------------------------------------------------------------------------------------------ Intercept 127.4246 185.907 0.685 0.494 -239.176 494.025 C(Q("urbanr (%)"))[T.(32.8, 55.2]] 211.0778 251.045 0.841 0.401 -283.973 706.128 C(Q("urbanr (%)"))[T.(55.2, 77.6]] 905.2163 236.449 3.828 0.000 438.949 1371.484 C(Q("urbanr (%)"))[T.(77.6, 100]] 2271.6658 262.912 8.640 0.000 1753.214 2790.117 ============================================================================== Omnibus: 196.152 Durbin-Watson: 2.055 Prob(Omnibus): 0.000 Jarque-Bera (JB): 4118.605 Skew: 3.766 Prob(JB): 0.00 Kurtosis: 23.741 Cond. No. 5.26 ==============================================================================
Warnings: [1] Standard Errors assume that the covariance matrix of the errors is correctly specified. Means for relectricperperson by urbanr status: relectricperperson urbanr (%) (10.31, 32.8] 127.424568 (32.8, 55.2] 338.502319 (55.2, 77.6] 1032.640869 (77.6, 100] 2399.090332
Standard deviations for relectricperperson by urbanr status: relectricperperson urbanr (%) (10.31, 32.8] 344.180664 (32.8, 55.2] 313.532440 (55.2, 77.6] 1103.424927 (77.6, 100] 2194.876709
MultiComparison summary: Multiple Comparison of Means - Tukey HSD,FWER=0.05 =============================================================== group1 group2 meandiff lower upper reject --------------------------------------------------------------- (10.31, 32.8] (32.8, 55.2] 211.0778 -439.3829 861.5385 False (10.31, 32.8] (55.2, 77.6] 905.2163 292.5745 1517.858 True (10.31, 32.8] (77.6, 100] 2271.6658 1590.4579 2952.8737 True (32.8, 55.2] (55.2, 77.6] 694.1385 115.8783 1272.3986 True (32.8, 55.2] (77.6, 100] 2060.588 1410.1273 2711.0487 True (55.2, 77.6] (77.6, 100] 1366.4495 753.8078 1979.0913 True ---------------------------------------------------------------
#############################################
Model Interpretation for ANOVA:
When examining the association between residential electricity consumption per person (quantitative response) and binned oil consumption per person (categorical explanatory), the ANOVA test revealed that among countries with the relevant data available, the subgroups of countries with intermediate oil consumption per person (3.08-6.13 and 6.13-9.18 litres) reported significantly greater consumption of residential electricity per person (m=6449.380371, s.d.=±4128.518555 and m=8362.568359) compared to the subgroups with the lowest oil consumption per person (m=830.4105, s.d.=± 1108.687744), F(3, 199) =35.72, p <0001.
On the other hand, when examining the association between residential electricity consumption per person (quantitative response) and binned urban population (categorical explanatory), the ANOVA test revealed that among countries with the relevant data available, the subgroups of countries with higher urban population (55.2-77.6 and 77.6-100%) reported significantly greater consumption of residential electricity per person (m=1032.640869, s.d.=±1103.424927 and m=2399.090332, s.d.=± 2194.876709) compared to the subgroups with lowest urban population (m=127.424568, s.d.=±344.180664), F(3, 199) =31.26, p <0001.
#############################################
Model Interpretation for post hoc ANOVA:
ANOVA revealed that among countries with the relevant data available, the oil consumption per person (collapsed into 4 ordered categories, which is the categorical explanatory variable) and residential electricity consumption per person (quantitative response variable) were significantly associated, F(6, 196)= 35.72, p=1.63e-18. Post hoc comparisons of mean residential electricity consumption by pairs of oil consumption per person categories revealed that other than the comparisons between the group with lowest and highest consumption (i.e. 0.02-3.08 and 9.18-12.23 litres) and the 2 intermediate groups (i.e. 3.08-6.13 and 6.13-9.18 litres), all other comparisons were statistically significant.
Additionally, ANOVA also revealed that among countries with the relevant data available, the urban population (collapsed into 4 ordered categories, which is the categorical explanatory variable) and residential electricity consumption per person (quantitative response variable) were significantly associated, F(6, 196)= 31.26, p=1.33e-16. Post hoc comparisons of mean residential electricity consumption by pairs of urban population categories revealed that other than the comparisons between the 2 subgroups with the lowest urban population (i.e. 10.3-32.8 and 32.8-55.2%), all other comparisons were statistically significant.
0 notes
Text
Terry Crews and his roles as a protective, attentive, hard-working, caring father in most of his iconic roles




#b99#terry jeffords#cotc#duane williams#cloudy with a chance of meatballs#everybody hates chris#julius rock#multicomparison#comparison appreciation#great parenting#trivia#my posts
154 notes
·
View notes
Text
Wanted to add a few examples other tumblr users have posted as well as my own.






















































“you can’t make a lawful good character interesting and enjoyable”:

398K notes
·
View notes
Text
Data Analysis Tools - Week 1
(gapminder dataset)
Model Interpretation for ANOVA:
Using income per person variable, I put the bottom 1/3, middle 1/3, and top 1/3 percentile into a 3,2,1 values, respectively. This variable is the income percentile group or perc_group.
When examining the association between employment rate and income percentile group, an Analysis of Variance (ANOVA) there is a significant difference between the top, middle, and bottom, with the mean and standard deviation for top percentile is a Mean=57.34% / s.d. ±7.51%, for the middle percentile is a Mean=55.22% / s.d. ±9.46%, and bottom percentile is a Mean=64.39% / s.d. ±11.53%, F score=13.66, p value = 3.27e-06. Since the p value is less than 0.05 there is a significant difference between the means and we reject the null hypothesis.
The degrees of freedom can be found in the OLS table as the DF model and DF residuals 2 and 163, respectively.
OLS Regression Results ============================================================================== Dep. Variable: employrate R-squared: 0.144 Model: OLS Adj. R-squared: 0.133 Method: Least Squares F-statistic: 13.66 Date: Mon, 04 Sep 2017 Prob (F-statistic): 3.27e-06 Time: 23:47:31 Log-Likelihood: -609.75 No. Observations: 166 AIC: 1226. Df Residuals: 163 BIC: 1235. Df Model: 2 Covariance Type: nonrobust ====================================================================================== coef std err t P>|t| [95.0% Conf. Int.] -------------------------------------------------------------------------------------- Intercept 57.3386 1.274 45.020 0.000 54.824 59.854 C(perc_group)[T.2] -2.1145 1.826 -1.158 0.249 -5.720 1.491 C(perc_group)[T.3] 7.0541 1.817 3.881 0.000 3.465 10.643 ============================================================================== Omnibus: 0.415 Durbin-Watson: 1.951 Prob(Omnibus): 0.813 Jarque-Bera (JB): 0.460 Skew: -0.118 Prob(JB): 0.794 Kurtosis: 2.894 Cond. No. 3.68 ==============================================================================
Warnings: [1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Model Interpretation of post hoc ANOVA results:
ANOVA analysis p value was significant, but to determine which of the means are significantly different I ran a Post hoc test, using ‘Tukey Honest Sigificant Difference’.
Multiple Comparison of Means - Tukey HSD,FWER=0.05 ============================================ group1 group2 meandiff lower upper reject -------------------------------------------- 1 2 -2.1145 -6.434 2.2049 False 1 3 7.0541 2.7549 11.3534 True 2 3 9.1687 4.8112 13.5261 True --------------------------------------------
When compared with employment rate, there is a difference between top and bottom income percentiles. There is also a difference between middle and bottom income percentiles.
Related Code:
import pandas as pd
data = pd.read_csv('GapminderData.csv')
# select the variables of interest df1 = data[['country','lifeexpectancy','employrate','femaleemployrate','DevelCountry','incomeperperson','perc_group']]
# new variable - earningpower (lifeexpectancy * incomeperperson) - this is not an average lifetime income but an indication of earning power potential over ones lifetime
df1['earningpower'] = df1['lifeexpectancy'] * df1['incomeperperson']
# new variable - unemployment (100 - 'employrate') - this is the percentage of the population who does not hold a job
df1['unemployment'] = 100 - df1['employrate']
# new variable - femaleunemployment (100 - 'femaleemployrate') - this is the percentage of females who does not hold a job
df1['femaleunemployment'] = 100 - df1['femaleemployrate']
# how many NAs do we have in employrate
print(df1['employrate'].value_counts(dropna=False))
# drop countries that does not have employrate variable
df1 = df1[pd.notnull(df1['employrate'])]
# did we remove the NAs in employrate variable?
print(df1['employrate'].value_counts(dropna=False))
# how many NAs do we have in lifeexpectancy
print(df1['lifeexpectancy'].value_counts(dropna=False))
# drop contries that have NAs in lifeexpenctancy
df1 = df1[pd.notnull(df1['lifeexpectancy'])]
# did we remove the NAs in lifeexpectancy
print(df1['lifeexpectancy'].value_counts(dropna=False))
df1.dropna(inplace=True)
#%%
df4 = df1[['employrate','perc_group']]
#%%
import statsmodels.formula.api as smf import statsmodels.stats.multicomp as multi
model = smf.ols(formula='employrate ~ C(perc_group)',data=df4) results = model.fit() print(results.summary())
#%%
df5=df4.groupby('perc_group').mean() df6=df4.groupby('perc_group').std()
print(df5,df6)
#%%
mc = multi.MultiComparison(df4['employrate'],df4['perc_group']) result2 = mc.tukeyhsd() print(result2.summary())
0 notes
Text


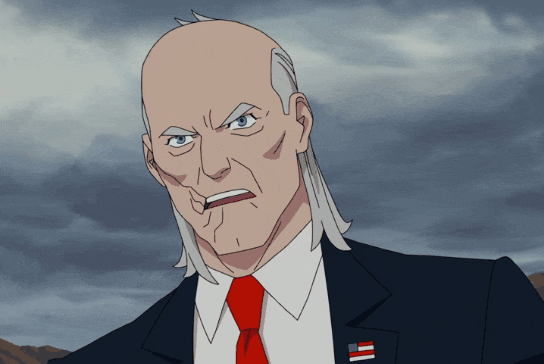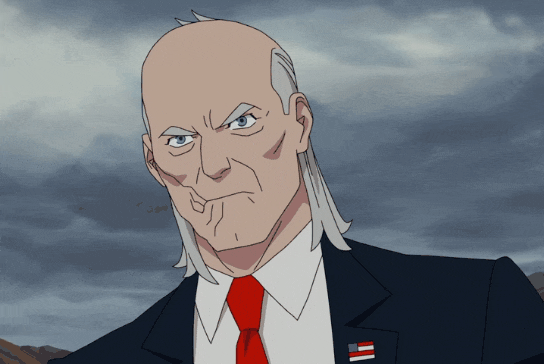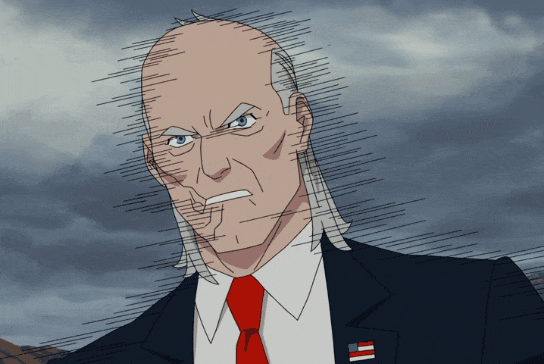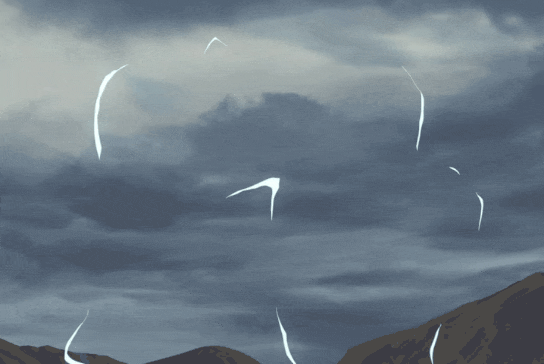
#dc#dcau#amanda waller#marvel#mcu#nick fury#invincible 2021#cecil stedman#multicomparison#mypolls#polls#my posts
13 notes
·
View notes
Photo
I would like a story exploring this “proof” with it being thoroughly debunked by using the above characters or rather, expies of them

collage of female characters that people have used to “prove” that there are too many masc women in media
#multimedia#multicomparison#comparison appreciation#fandom critical#feminism#my add on#story idea#mine#expy idea
96K notes
·
View notes
Text
For some reason, I think this trio of blue-themed, intelligent, socially awkward but still heroic young women who faced unfair ostracization from others would be great friends



#lol arcane#lol caitlyn#star trek lower decks#t'lyn#dc#dcu#nina mazursky#colors#multicomparison#comparison appreciation#team idea#my posts#myteam
16 notes
·
View notes