#eukaryotic cells
Explore tagged Tumblr posts
Text
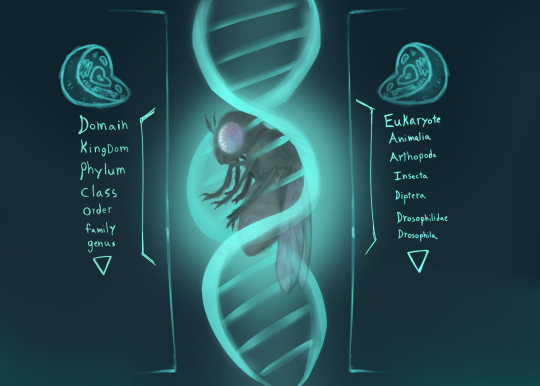
fruitfly
#insects#bugs#art#original stuff#silly#fruit fly#animal classification#genetics#eukaryotic cells#arthropods
11 notes
·
View notes
Text
Development of Cancer

Patreon
#studyblr#notes#my notes#medblr#cell biology#eukaryotes#eukaryotic cells#eukaryotic cell biology#cell biology notes#cell bio#cell bio notes#cytology#cytology notes#cell chemistry#cell biochemistry#eukaryotic cytology#cell functions#functions of cells#eukaryotes notes#study of cells#cell basics#science#scienceblr#life science#health science#cancer#cancer biology
9 notes
·
View notes
Text

page 199 - do these plants look like my genitals? do my genitals look like these plants? are these funguses? at this point, are my genitals? or are they just covered in thousands of tiny mushrooms?
It's all well and good to have a fun guy at your party but I'll take a bunch of fun Guses any day of the week. Those people can party.
#zoology#zoologist#diagram showing major parts of human male and female reproductive systems as seen in imaginary median sections#colon#alien plants#alien#biome#primordial#eukaryotes#eukaryota#eukaryotic cells#mushrooms#fungus#mushroom#magic#ovary#fallopian tubes#testes
4 notes
·
View notes
Text
The first prokaryotes, or microscopic single-celled organisms without a distinct nucleus, likely emerged by 3.9 billion years ago but these Eukaryotic fossils appear 70 million years earlier than scientists previously thought multicellular life arose.
2 notes
·
View notes
Text
Even the Oldest Eukaryote Fossils Show Dazzling Diversity and Complexity - Technology Org
New Post has been published on https://thedigitalinsider.com/even-the-oldest-eukaryote-fossils-show-dazzling-diversity-and-complexity-technology-org/
Even the Oldest Eukaryote Fossils Show Dazzling Diversity and Complexity - Technology Org
The sun has just set on a quiet mudflat in Australia’s Northern Territory; it’ll set again in another 19 hours. A young moon looms large over the desolate landscape. No animals scurry in the waning light. No leaves rustle in the breeze. No lichens encrust the exposed rock. The only hint of life is some scum in a few puddles and ponds. And among it lives a diverse microbial community of our ancient ancestors.
A soft summer evening in the Paleoproterozoic, as envisioned by DALL-E. Image credit: DALL-E, prompt by Harrison Tasoff.
In a new account of exquisitely preserved microfossils, researchers at UC Santa Barbara and McGill University revealed that eukaryotic organisms had already evolved into a diverse array of forms even 1.64 billion years ago.
The paper, published in the journal Papers in Paleontology, recounts an assemblage of eukaryotic fossils from an era early in the group’s evolutionary history. The authors describe four new taxa, as well as evidence of several advanced characteristics already present in these early eukaryotes.
“These are among the oldest eukaryotes that have ever been discovered,” explained lead author Leigh Anne Riedman, an assistant researcher in UCSB’s Department of Earth Science. “Yet, even in these first records we’re seeing a lot of diversity.”
Eukarya forms one of the major domains of life, encompassing the plant, animal and fungi clades, as well as all other groups whose cells have a membrane-bound nucleus, like protists and seaweeds.
Many scientists had thought early eukaryotes were all fairly similar during the late Paleoproterozoic, and that diversification took place around 800 million years ago. But Riedman and her co-authors found fossils of a delightfully diverse, and complex, cast of characters in rock nearly twice as old.
Limbunyasphaera operculata is a new species that shows a small door opening into the cell. Photo Credit: Riedman et al.
Scientists knew from previous studies that eukaryotes had evolved by this time, but their diversity in this era was poorly understood. So Riedman headed to the Outback in late 2019. Within one week, she had collected about 430 samples from eight cores drilled by a prospecting company; they now reside in the library of the Northern Territory Geological Survey. The two cores used for this study spanned roughly 500 meters of stratigraphy, or 133 million years, with around 15 million years of significant deposition.
Riedman returned to the United States with shale and mudstone: remnants of an ancient coastal ecosystem that alternated between shallow, subtidal mudflats and coastal lagoons. A dip in hydrofluoric acid dissolved the matrix rock, concentrating the precious microfossils which she then analyzed under the microscope.
“We were hoping to find species with interesting and different characteristics to their cell walls,” Riedman said. She hoped that these features could shed light on what was happening within the cells during this time period. Reaching any conclusions about the cellular interior would require a great deal of sleuthing, though, since the fossils preserve only the exterior of the cells.
The researchers were surprised by the diversity and complexity preserved in these fossils. They recorded 26 taxa, including 10 previously undescribed species. The team found indirect evidence of cytoskeletons, as well as platy structures that suggest the presence of internal vesicles in which the plates were formed — perhaps ancestral to Golgi bodies, present in modern eukaryotic cells. Other microbes had cell walls made of bound fibers, similarly suggestive of the presence of a complex cytoskeleton.
The authors also found cells with a tiny trapdoor, evidence of a degree of sophistication. Some microbes can form a cyst to wait out unfavorable environmental conditions. In order to emerge, they need to be able to etch an opening in their protective shell.
Making this door is a specialized process. “If you’re going to produce an enzyme that dissolves your cell wall, you need to be really careful about how you use that enzyme,” Riedman said. “So in one of the earliest records of eukaryotes, we’re seeing some pretty impressive levels of complexity.”
Many people in the field had thought this ability emerged later, and the evidence for it in this assemblage further emphasizes how diverse and advanced eukaryotes were even at this early juncture.
“The assumption has always been that this is around the time that eukaryotes appeared. And now we think that people just haven’t explored older rocks,” said co-author Susannah Porter, an Earth science professor at UC Santa Barbara.
This paper is part of a larger project investigating early eukaryote evolution. Riedman and Porter want to know in what environments early eukaryotes were diversifying, why they were there, when they migrated to other places, and what adaptations they needed in order to fill those new niches.
A big part of this effort involves understanding when different characteristics of eukaryotes first arose. For instance, the authors are quite interested to learn whether these organisms were adapted to oxygenated or anoxic environments.
The former would suggest that they had an aerobic metabolism, and possibly mitochondria. Every modern eukaryote that’s been found descends from ancestors that possessed mitochondria. This suggests that eukaryotes acquired the organelle very early on, and that it provided a significant advantage.
Riedman and Porter are currently working on a fresh account of eukaryote diversity through time. They’ve also collected even older samples from Western Australia and Minnesota. Meanwhile, their geochemist collaborators at McGill are spearheading a study on oxygen levels and preferred eukaryote habitats, aspects that could shed light on their evolution.
“These results are a directive to go look for older material, older eukaryotes, because this is clearly not the beginning of eukaryotes on Earth,” Riedman said.
Source: UCSB
#Animals#Australia#billion#Biotechnology news#cell#Cells#Chemistry & materials science news#Community#complexity#dall-e#deal#diversity#domains#earth#Environmental#enzyme#eukaryotic cells#Evolution#Explained#Features#fibers#form#Forms#Fossils#fungi#Genetic engineering news#History#how#it#Landscape
2 notes
·
View notes
Text
OOoooh these are GORGEOUS!


Eukaryotic cell gang!! We love women in STEM.
The organelles of the cells have been translated into human anatomy, so the nucleus is the brain, the vacuole function as the lungs, and the mitochondria is the heart since it’s the… you already know, I don’t have to say it ;)
121K notes
·
View notes
Text
She eukaryote on my helminth tell I nematode
0 notes
Text



#cat#homes#nature#architechture#Space#Universe#furry#Exotic#Projects#3dgraphics#Coating#Appliances#Grooming#eukaryotic cells
1 note
·
View note
Video
Protein Modules In Signal Transduction: A Lecture by National Library of Medicine Via Flickr: Series Title(s): NIH director's Wednesday afternoon lecture series Contributor(s): Pawson, T. National Institutes of Health (U.S.). Medical Arts and Photography Branch. Publication: [Bethesda, Md. : Medical Arts and Photography Branch, National Institutes of Health, 1998] Language(s): English Format: Still image Subject(s): Signal Transduction, Gene Expression Regulation -- physiology, Eukaryotic Cells -- physiology Genre(s): Posters Abstract: Black, orange, and red poster with a visual representing signal transduction. Details of the event are also given. Extent: 1 photomechanical print (poster) : 71 x 41 cm. Technique: color NLM Unique ID: 101456172 NLM Image ID: C02937 Permanent Link: resource.nlm.nih.gov/101456172
#photomechanical print#poster#print#Medical Arts and Photography Branch#NIH director's Wednesday afternoon lecture series#Gene Expression Regulation#physiology#Eukaryotic Cells#Protein Modules#Signal Transduction#lecture#Still Image#Public Domain#Free Images#Prints and Photographs#National Library of Medicine#NLM#IHM#National Institutes of Health#NIH#Archives of Medicine#NLM Digital Collection#flickr
0 notes
Text
When Nuclei Awakes
A Journey into the Depths of Cellular ConsciousnessIn a moment of profound awakening, it’s said that every fibre of our being tunes into a profound truth, a realization that resonates through every cell, molecule, and atom within us. The human body, a vast universe of about 37.2 trillion cells, becomes a mirror reflecting this profound awareness. Among these, about 30 trillion are eukaryotic…

View On WordPress
#Akashic Field#Cellular Consciousness#Eukaryotic Cells#Gluons#Interconnectedness#Nuclear Physics#Quantum Biology#spiritual awakening#Subatomic Particles#Zero Point Energy
1 note
·
View note
Text
I don't know the context, but I just want to say that sperm has got to be one of the worst cells in the human body. It's got some good features. The acrosome is neat, I can applaud the helical mitochondrion. None of these features outweigh the travesty that is the eukaryotic flagella!
Needlessly complex systems of dyneins, and for what! Some bending??? Minor undulations? This can be excused in the cilia but for a flagellum it's just pointless. Look at the prokaryotic flagella:

(Nicholas et al. 2021)
Look at it. Look! Bathe in it's beauty, it's simplicity, it's elegance!
V-ATPase is one of if not the greatest enzymes! And it's situated right at the base of the procaryotic flagella. A flagellum that truly rotates! Imagine that! Eukaryotes couldn't even dream of such a thing. They disgrace biology with their crude imitations of rotation, their facsimile of a motor. They could never know beauty, and every instance of a eukaryotic flagellum simply disgusts me.
And think! of where we discovered the complex and spectacular structure of ATP Synthase, and it's relative, V-ATPase! Think of where we gleamed the emergent and elegant prokaryotic flagella structure in the first place! It of course came from The Model Organism, Escherichia Coli. An organism that makes its ever hallowed movement through extracellular space using a complex of prokaryotic flagella. The ability to switch direction of rotation aiding in the fluid and graceful locomotion of its rod shaped gram negative cell body.
Works cited: Nicholas J. Matzke, Angela Lin, Micaella Stone, Matthew A. B. Baker (2021). Homology between the flagellar export apparatus and ATP synthase: evidence from synteny predating the Last Universal Common Ancestor. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.01.01.425057v2


New nightmare just dropped

I do not like the Spworm. 😶🌫️🪱
#biology#rant#prokaryotes#eukaryotic cells#nsft eukaryotic flagella#cw: eukaryotic flagella#jen rants
120 notes
·
View notes
Text
Major Eukaryotic DNA Polymerases

Patreon
#studyblr#notes#medblr#medical notes#med notes#polymerases#dna#genetics#genetics notes#biology#biology notes#bio#bio notes#biochemistry#dna polymerases#eukaryotes#eukaryotic cells#eukaryotic genetics#eukaryotic polymerases#eukaryotic dna#eukaryotic dna polymerases#primers#dna replication#nuclear genetics#nuclear genome#genome#genomes#inheritance#genetic inheritance
3 notes
·
View notes
Text

Prisoner playing the piano, since it is quite prominent in her part of the soundtrack. Also an excellent excuse to draw dapper Prisoner.
#slay the princess#stp princess#stp prisoner#piano#my art#cw: decapitation#with apologies to pianokind#i neither play nor understand pianos#when looking at the reference my mind started going to diagrams of the eukaryotic cell#'ah this is the piano's endoplasmatic reticulum#that's the cytoskeleton#and those bits are the golgi apparatus'#i figured out after a while it's probably because i'm trash at cell biology#so it was like 'okay i used to have to draw (diagrams of) complicated things i didn't understand in university too#spool up those same neurons'
52 notes
·
View notes
Text
This generation is so soft. "oh no, I can't eat that. what if it has germs on it and I get sick?"
In the time of our glorious ancestors, people were adventurers. You would go up to any random germ and press it into an indentation in your skin until it went straight into your body cavity! Who knows what happens next. Maybe you digest it and gain some nutrition. Maybe it survives inside you and gives you the ability to photosynthesize! People were able to achieve greatness because they weren't cowards about what they put in their bodies.
8 notes
·
View notes
Photo
I might sound like a basic bitch, but the mitochondria, but not just because 'its the powerhouse of the cell'. Because it's the site of oxidative and nonoxidative phosphorylation, because it's the only thing in our body that literally has its own unique DNA different from the rest of the body. It's my favorite because the ongoing theory of how it came to be in eukaryotic cells is that it was a separate microorganism/bacteria that was enveloped into the cell and they had mutually beneficial relationship (endosymbyosis) and it just stayed?! Like a permentant live in bestie?! Mitochondria said, hey if you protect me I'll give you a shit ton of adenosine triphosphate and now she's a legend. It's the home of bioenergetics, which people spend millions of dollars and their whole careers studying. We literally produce OUR OWN BODY WEIGHT in ATP per day thanks to the mitochondria all over our body which is literally fucking insane y'all because those little bitches are tiny af.
Thank you for reading my kinesiology student word vomit about my favorite little guys :)
Below are some links to free textbook pages if you are at all curious to read more.
If even 1 person learns a new fact from this and thinks this is cool I think I'll die from happiness. Comment or dm me if you like it plz 🥹

#science#ted talks#bioenergetics#biology#human anatomy#anatomy#microbiology#mitochondria#organelles#eukaryotic cells#cells#kinesiology#sports science
186 notes
·
View notes
Text
Illuminating a critical step in initiating DNA replication in eukaryotes - Technology Org
New Post has been published on https://thedigitalinsider.com/illuminating-a-critical-step-in-initiating-dna-replication-in-eukaryotes-technology-org/
Illuminating a critical step in initiating DNA replication in eukaryotes - Technology Org
Brandt Eichman and Walter Chazin, professors of biochemistry, have worked together to better understand how DNA replication is initiated in eukaryotes. Using Vanderbilt’s state-of-the-art instrumentation in the Center for Structural Biology’s Cryo-Electron Microscopy Facility, Eichman, Chazin, and their colleagues provided detailed visualizations of a multi-functional protein in action, which sheds light on how DNA replication is initiated in humans.
Cryo-EM structures of polα–primase reveal a remarkable range of motion between two sub-complexes.
Eichman and Chazin shared reflections on this research, newly published in Nature Structural & Molecular Biology:
What issue does your research address?
We are interested in the molecular details of human DNA replication, one of the most fundamental processes of life; it is repeated millions of times each day as we make new cells. The new copies of DNA are synthesized by polymerases, which read the sequence of an existing DNA strand one nucleotide at a time and add the complementary nucleotide to the nascent DNA strand. Specific polymerases perform the bulk of DNA synthesis but cannot function without first having a short “primer” segment of the new strand.
This work addresses the molecular mechanisms of DNA polymerase α–primase (polα–primase), the enzyme responsible for synthesizing the primers. Polα–primase is an essential enzyme as it is the only polymerase that can initiate DNA synthesis by generating the primers that the other polymerases need to duplicate the genome.
Despite polα–primase being the first human polymerase discovered, the way it synthesizes very specific lengths of RNA and DNA in a single strand remained unclear for more than fifty years. How does it know that it has synthesized a specific number ofnucleotides of RNA before transitioning to DNA synthesis? How does it transition between the two modes? How does it know that it has synthesized a certain number of nucleotides of DNA before stopping?
Understanding the mechanisms behind polα–primase’s ability to “count” the length of the RNA and DNA segments of the primer is important because primers must be kept to a very short length, as they contain RNA in the new DNA strand and the DNA synthesized by polα is littered with mutations. Thus, the primers would be highly detrimental to the cell if they became a substantial part of the new DNA strand that persisted in the genome after replication.
To answer these outstanding questions, we used cryo-electron microscopy to capture snapshots of this multi-functional protein at various stages as it generates a primer. The high-resolution structures we determined illuminated the mechanisms of RNA and DNA counting by polα–primase. They also provide a starting point for design of novel small molecule modulators of polα–primase function that would provide new ways to investigate DNA replication in cells.
What was unique about your approach to the research?
The Eichman and Chazin labs have collaborated for many years to understand how polα–primase works. We visualized some of the first structures of polα–primase bound to nucleic acid substrates. It was the highly strategic design of primer/template substrates that allowed our team to “trap” the enzyme at several specific points along the pathway to synthesizing the primer. Importantly, this research was made possible by access to the state-of-the-art instrumentation in the CSB Cryo-Electron Microscopy Facility.
What were your findings?
Our data directly show that polα–primase holds on to one end of the primer throughout all stages of synthesis. This observation is critical to understanding how the initial RNA-primed template is handed off from the primase active site in one subunit (where RNA synthesis occurs) to the DNA polymerase active site in another subunit (where DNA synthesis occurs). The sustained attachment also serves to increase polα–primase’s ability to remain bound to the template and to regulate both RNA and DNA composition. Importantly, the detailed analysis of the structures revealed how flexibility within this four-subunit complex is critical to being able to synthesize the primer strand across two active sites.
In addition, our research suggests that termination of DNA synthesis is facilitated by reduction polα and primase affinities for the template as more DNA is synthesized.
What do you hope will be achieved with the research results?
We hope our research findings will illuminate to the field a more complete understanding of replication initiation and contribute to the growing understanding that complex molecular machinery requires flexibility and dynamics to function. The inherent flexibility within this complex, multi-subunit polymerase is essential to primer synthesis and to its ability to dynamically interact with multiple other enzymes present in the replisome (for the handoff of the primer to the replicative polymerase for bulk DNA synthesis, for example).
We also hope that this work will lead to a better understanding of how current polα–primase inhibitors work and more broadly pave the way for future designs of small molecule modulators to serve as tools for studying DNA replication in cells. Tool compounds of this type can also be used to evaluate the therapeutic potential of targeting specific replication proteins with roles in diseases of genome instability.
Source: Vanderbilt University
You can offer your link to a page which is relevant to the topic of this post.
#amp#Analysis#approach#Art#biochemistry#Biology#Biotechnology news#Capture#cell#Cells#Composition#cryo-electron microscopy#data#Design#details#Diseases#DNA#DNA replication#dynamics#electron#enzyme#enzymes#eukaryotic cells#Fundamental#Future#Genetic engineering news#genome#High-Resolution#how#human
0 notes
