#tnfa
Explore tagged Tumblr posts
Text
The Doctor and The Nurse



!!! SPOILER ALERT!!! Continue at your own risk...
Welcome to the first meeting of the "The Nurse Fangirl Association" (short TNFA).
I watched "The Robot Revolution" yesterday. Yep. They got me. Belinda Chandra, aka "The Nurse", is glorious!
I love her energy. Very Donna Noble vibes, as my bestie put it. I love Ruby Sunday just as much, but for different reasons, them being hugely different characters.
Belinda doesn't take shit from nobody and I have the feeling we will have lots of fun with her...
With Ncuti, they keep following the path they started with the 1st season, him being this emontional and I for one am totally here for it. The way he bonded to Sasha 55 after 6 months and how it broke his heart when she passed.
*sigh*
The robot design was so cute. The cute ray-guns and how they held them. 😍 Okay, yes, they were bad most of the episode and killed people, but still...forgive me my enthusiasm for "silly villain design".
Oh, almost forgot...HOW ADORABLE WAS NCUTI'S OUTFIT AT THE BEGINNING!?! HELLO?!? He is so over-the-top with his wardrobe. I. Love. that. so. much.
It just adds to the overally excitement I am feeling right now towards the upcoming episodes. This was really more than just a promising start...
#doctor who#15th doctor#the robot revolution#belinda chandra#the nurse#ncuti gatwa#varada sethu#sasha 55#belindachandra#planet belindachandra#the ai generator
16 notes
·
View notes
Text
Ranking cells of the immune systems
Eosinophil - I have asthma -10000/10
Basophil - who? 5/10
Neutrophil - the farmers and peasants that got roped into this crusade, make up for half of the army but die off quickly. have more weapons than they should and are in larger numbers than they should be. the skaven of the immune system. 8/10
Macrophage (resident and patroller) - like neutrophils, they carry this entire body on their back. Who kickstarts the major immune reaction? Whos producing IL6 and TNFa??? Thats right bitch 10/10
Monocyte - baby macrophage 10/10
Lymphocyte T CD4 - squire. Trying their best, kind of cute .... kind of very nosy 7/10
Lymphocyte T CD8 - paladin class of the immune system. 9/10
Lymphocyte B - half alchemists 5/10
Plasm cell - full metal alchemists 10/10
NK cells and all their derivatives - godzilla let loose 10000/10
11 notes
·
View notes
Text
there's a lot of data floating around about antidepressants (specifically tricyclics and SSRIs) as a decently effective form of treatment for certain gastrointestinal symptoms, and part of me has always been a little resistant to the idea because it risks coming across as an "actually it was all in your head" thing—and tbh frequently does come across that way lol)—but if you cut through the vague air of dismissiveness or whatever there's still good reason to believe it's legit since (1) we're pretty confident the ~gut-brain axis~ exists even if we're not sure exactly how or why and (2) anything that affects the brain is bound to affect the rest of the body since, like, the brain is what's piloting the whole machine. there's a reason anti-emetics like zofran target the brain directly and not your stomach. and literally anyone with GI problems can tell you that worrying about those problems is almost guaranteed to make them worse.
BUT, APPARENTLY, wellbutrin (an SNRI) takes that shit to the next level by directly reducing TNFa levels in your gut. or allegedly does anyway, at least a little bit. and for the uninitiated, TNFa is an inflammatory protein that does useful stuff in your body but can get a little carried away with it sometimes (i.e., if you have an autoimmune disease); it's not the only culprit for intestinal inflammation, but it's one of the most common ones, so some of the most common drugs used to treat crohn's and colitis are "TNF inhibitors" like remicade and humira and enbrel. except where all of those are highly specialized artificial antibodies or genetically engineered shit that has to be grown painstakingly in a lab, wellbutrin is just a random pill. and obviously its effect on TNFa won't be as strong as an intravenous medication, and it's not the only random pill to affect TNF levels (apparently thalidomide also does lol) but I'm still like what the fuck that's crazy. and also wondering if I would've developed crohn's symptoms earlier if I hadn't been on wellbutrin in my teens and early 20s...
24 notes
·
View notes
Text
The TNFA stock touches 52-week less than $ 0.32 Aid the challenges
The TNFA stock touches 52-week less than $ 0.32 Aid the challenges Source link

View On WordPress
0 notes
Text
The TNFA stock touches 52-week less than $ 0.32 Aid the challenges
The TNFA stock touches 52-week less than $ 0.32 Aid the challenges Source link

View On WordPress
0 notes
Text
Stephana
Me da imagenes (TNFa) y no me explica nadie que debo poner o no poner en la tesis. Por ende tengo que interpretar yo solo los resultados.
Le solicito resultados que van en mi tesis y se olvida de enviarlos. Luego me culpan por no poner la grafica que deberia ir ahi.
Revisa la tesis al ultimo momento, solo para eliminar, graficas que me tomaron tiempo en hacer. Con sus respectivos analisis estadisticos.
Me culpa de no escribir pesimos resultados pero no se tomo el tiempo de explicarme que debo poner en las partes que no son tan evidentes (TNFa o interpretacion de Alamar blu).
Revisa la tesis al ultimo, y quita dos o tres secciones que Maxime me ha dicho que poner.
Habla fuerte y exige pero no sabe reconocer los limites del respeto
Maxime
Maxime me pide que ponga stats pero no me dice si necesita numeros. Luego me escribe para pedirme que ponga los simbolos. Luego que remueva los numeros.
Me obliga a poner resultados incompletos de MTT assay y solo hasta reconocer que no son significativos me autoriza usar los de Alamar Blue.
Nunca se muestra abierto a recibir ayuda cuando le ofrezco ayuda. Me hace ver como que soy un estudiante flojo pero nunca me pide nada en el laboratorio. Vengo varios dias solo a sentarme o a realizar solo trabajos menores en el lab.
Me sugiere inventar informacion de graficas.
Clarissa
Habla fuerte y exige pero no sabe reconocer los limites del respeto.
0 notes
Text
Needing to dilute samples for analysis is so stupid what do you MEAN my cells produced TOO MUCH tnfa for your stinky elisa standard curve??? I fail to see how that's MY problem no I don't want to have to dilute 1:50 honestly my cells should be REWARDED for being so good and reactive and you come into MY HOUSE AND- *I am shot dead by BioTechne technical support*
1 note
·
View note
Text
Recombinant Canine TNF-alpha
Recombinant Canine TNF-alpha Catalog number: B2017868 Lot number: Batch Dependent Expiration Date: Batch dependent Amount: 1 μg Molecular Weight or Concentration: ~17.3 kDa Supplied as: Powder Applications: a molecular tool for various biochemical applications Storage: -20°C Keywords: TNFA, cTNF, cachectin, tumor necrosis factor ligand superfamily member 2 Grade: Biotechnology grade. All products…
0 notes
Text
Nutrients, Vol. 16, Pages 1929: Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice
Background: High-fat diets cause gut dysbiosis and promote triglyceride accumulation, obesity, gut permeability changes, inflammation, and insulin resistance. Both cocoa butter and fish oil are considered to be a part of healthy diets. However, their differential effects on gut microbiome perturbations in mice fed high concentrations of these fats, in the absence of sucrose, remains to be elucidated. The aim of the study was to test whether the sucrose-free cocoa butter-based high-fat diet (C-HFD) feeding in mice leads to gut dysbiosis that associates with a pathologic phenotype marked by hepatic steatosis, low-grade inflammation, perturbed glucose homeostasis, and insulin resistance, compared with control mice fed the fish oil based high-fat diet (F-HFD). Results: C57BL/6 mice (5–6 mice/group) were fed two types of high fat diets (C-HFD and F-HFD) for 24 weeks. No significant difference was found in the liver weight or total body weight between the two groups. The 16S #rRNA sequencing of gut bacterial samples displayed gut dysbiosis in C-HFD group, with differentially-altered microbial diversity or relative abundances. Bacteroidetes, Firmicutes, and Proteobacteria were highly abundant in C-HFD group, while the Verrucomicrobia, Saccharibacteria (TM7), Actinobacteria, and Tenericutes were more abundant in F-HFD group. Other taxa in C-HFD group included the Bacteroides, Odoribacter, Sutterella, Firmicutes bacterium (AF12), Anaeroplasma, Roseburia, and Parabacteroides distasonis. An increased Firmicutes/Bacteroidetes (F/B) ratio in C-HFD group, compared with F-HFD group, indicated the gut dysbiosis. These gut bacterial changes in C-HFD group had predicted associations with fatty liver disease and with lipogenic, inflammatory, glucose metabolic, and insulin signaling pathways. Consistent with its microbiome shift, the C-HFD group showed hepatic inflammation and steatosis, high fasting blood glucose, insulin resistance, increased hepatic de novo lipogenesis (Acetyl CoA carboxylases 1 (Acaca), Fatty acid synthase (Fasn), Stearoyl-CoA desaturase-1 (Scd1), Elongation of long-chain fatty acids family member 6 (Elovl6), Peroxisome proliferator-activated receptor-gamma (Pparg) and cholesterol synthesis (β-(hydroxy β-methylglutaryl-CoA reductase (Hmgcr). Non-significant differences were observed regarding fatty acid uptake (Cluster of differentiation 36 (CD36), Fatty acid binding protein-1 (Fabp1) and efflux (ATP-binding cassette G1 (Abcg1), Microsomal TG transfer protein (Mttp) in C-HFD group, compared with F-HFD group. The C-HFD group also displayed increased gene expression of inflammatory markers including Tumor necrosis factor alpha (Tnfa), C-C motif chemokine ligand 2 (Ccl2), and Interleukin-12 (Il12), as well as a tendency for liver fibrosis. Conclusion: These findings suggest that the sucrose-free C-HFD feeding in mice induces gut dysbiosis which associates with liver inflammation, steatosis, glucose intolerance and insulin resistance. https://www.mdpi.com/2072-6643/16/12/1929?utm_source=dlvr.it&utm_medium=tumblr
0 notes
Link
0 notes
Text
I’m getting my crp drawn on Friday and I’m holding my NSAIDs so it doesn’t skew the results but. Now I can’t sleep. I’m going to be in fucking agony the next few days. Just confirming my self diagnosis of seronegative axial spondylarthritis because I meet literally all the clinical features and have imaging to back it up(haha back). But no doctor has ever diagnosed me with anything besides “back hurty discs fucked” and regular osteoarthritis which, ouch, I’m 24(and was diagnosed at 19 😬😬😬). Idk I know it doesn’t matter because the treatment is NSAIDS which I take in kidney frying levels or TNFa antagonists, and I really don’t want to knock out my immune system when I work around sick people all the time. Augh I wish I could just cut open my back, scrub my spine with a scotch Brite pad, and snap everything back into place. Hell on earth
1 note
·
View note
Text
Cytokines and systemic lupus erythematosus
SLE is an autoimmune rheumatic disease that principally affects women in the childbearing years.1 The prevalence of SLE varies between ethnic groups, being approximately 1:250 among black women, 1:1000 in Chinese women and 1:4300 in white women. SLE affects the joints, skin and blood in over 80% of patients and the kidneys, central nervous system and cardiopulmonary system in 30%–50% of patients. Between 10% and 30% of patients have anticardiolipin antibodies that are associated with arterial and venous thrombosis. The majority of patients demonstrate systemic manifestations, which may include fatigue, malaise, fever, anorexia, nausea and weight loss.
Many cytokines have been implicated in regulating disease activity and in the involvement of different organs in patients with SLE. This article reviews both the role of individual cytokines and discusses possible mechanisms of cytokine action in individual organs.
0 notes
Text
Home
What Is Meant By TNF Alpha Elisa Kit Antibody?
The TNF alpha is a multifunctional proinflammatory cytokine, which belongs to the super family of tumor necrosis factor (TNF). The main functions of TNF alpha Elisa kit antibody are including its prime function in an immune response to the viral, bacterial, parasitic along with certain fungal infections as well as additionally its role in a necrosis of the particular tumors. The TNF alpha ELISA is an immune assay kit for the quantitative measurement of tumor necrosis factor alpha (TNFA) in the plasma, lysate samples and human serum and cell culture supernatants.Click here for more info https://www.bosterbio.com/
Exactly How Does TNF Elisa Works?
The quantikine human TNF alpha immune assay is a 3.5 to 4.5 hr solid phase Elisa made to determine the human TNF alpha in serum, cell culture supernates and plasma and so forth. It really includes E coli-delivered recombinant human TNF alpha in addition to antibodies increased against this protein. It has actually also been shown to completely quantitate the recombinant factor. According to the research, it has been shown that every immune assay can represent its drawbacks as well as benefits, only a few assays are appropriate for the measurement of TNF alpha in the clinical laboratories as well as exhibit that so far away the extent of TNF alpha in the human blood is still not yet synchronized.
The human TNF alpha Elisa kit is a solitary wash ninety min sandwich Elisa that specially made for the quantitative quantity of TNF alpha protein in the plasma, human serum and culture media. It additionally uses an exclusive simple step Elisa technology and quantitates the human TNF alpha with 4.32 pg/ml compassion. The Elisa technology really employs capture the antibodies that are conjugated to an affinity age, which is identified by the monoclonal antibody utilized to coat the basic step Elisa plates. This technique to sandwich Elisa allows the creation of antibody analyte sandwich difficult in one step and significantly minimizing the assay time.
Major Benefits Of Elisa Technology
The major advantages of basic action Elisa technology are consisting of:
- Entirely validated in biological samples
- Single wash protocol reduces assay time to 90 minutes or less
- 96 wells plate breakable into 12x8 wells strips
- High specificity, sensitivity and also reproducibility from the leading antibodies
Test Concept Of TNF Alpha Elisa Kit

The Elisa kit really utilizes a sandwich Elisa concept. This Micro Elisa plate offered in this kit has been pre-coated with an antibody certain to human TNF alpha. Its samples or standards are consisted of tomicro Elisa plate wells as well as additionally blended with the particular antibody. The TNF alpha Elisa kit is majorly triggered by the endotoxins, viral infections, lipopolysaccharides or any other bacterial parts by DNA damage, tissue injury as well as also by TNF alpha as well as IL-1, PDGF. It is generally expressed in neutrophils, macrophages, endothelial cells, NK-cells, mast cells, monocytes and activated lymphocytes. It additionally encourages the systematic or local inflammation along with urges the severe stage response. The dysregulation of TNF alpha is included in several diseases. To know more regarding this Elisa kit, you can click here to find out more.
1 note
·
View note
Text
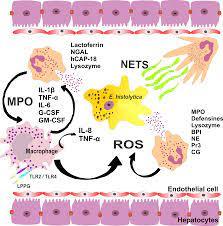
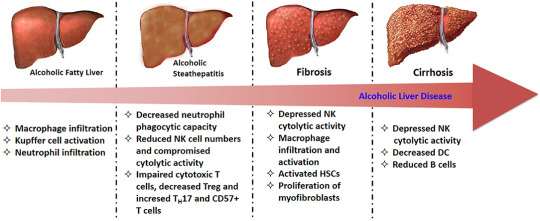
Alcoholic Liver Disease
Pathogenesis
Stage One
Ethanol is metabolised in the liver to acetaldehyde by alcohol dehydrogenase
which is in turn metabolised to acetate by aldehyde dehydrogenase.
As acetaldehyde is formed, NAD is metabolised to NADH as a cofactor in this reaction.
This NADH inhibits the actions of isocitrate dehydrogenase and alpha ketoglutarate and fatty acid oxidation, meaning it favours fatty acid synthesis.
Fat builds up in vacuoles of hepatocytes and mitochondria begin to dysfunction under oxidative stress, creating reactive oxygen species.
This fatty stage is known as steatosis and is stage one of alcoholic liver disease. It is reversible.
Stage Two
Inflammatory cytokines such as IL-6 and TNFa influx in, causing inflammation.
Water begins to accumulate inside hepatocytes and they begin to balloon. This causes significant cholestasis.
The inflammatory cytokines stimulate hepatocytes to produce collagen, which can begin to form fibrosis.
This collagen can be pericellular or around central veins, forming collagen bridges. This is a precursor for cirrhosis.
Stage Three
As the fibrosis progresses, hepatocytes begin to die and cirrhosis occurs.
This is end-stage alcoholic liver disease and can lead to liver failure and death.
Liver function tests
In stage 1; all LFTs will be raised. AST and ALT will be raised by approximately 5-10x the upper reference limit except albumin. They will both usually be below 300IU/L. AST will be raised above ALT; this is different to other hepatic conditions such as cholestasis, tumours, hepatitis, where ALT will be above AST.
In stage 2; ALP and bilirubin will increase significantly as cholestasis occurs and blocks bile duct. This will usually be above 300IU/L, which is different from filtrative diseases, where ALP is usually raised to 200-250IU/L. AST and ALT may increase further.
In stage 3; AST and ALT are between 10-100x the upper reference limit, suggesting significant cirrhosis and damage to cells. AST will remain above ALT. GGT will be raised significantly, which is common with alcoholic liver disease. Albumin will be decreased, suggestive of total liver failure. In other hepatic conditions, liver damage usually does not reach this stage and so albumin stays within the range.
#medicine#biomed#biomedicine#notes#medblr#studyblr#sciblr#premed#nursing#biology#human biology#med#biomedical science#science#liver#liver disease#alcoholic liver disease#ald#pathology#2#3#lft#clinical#hepatic#hepatic system
82 notes
·
View notes
Text
Infliximab


Common Brand Names: Remicade
Therapeutic Class: Monoclonal antibody, TNFa-blocking agent
Common Injectable Dosage Forms:
Lyophilized Powder for Injection: 100 mg/vial. Store under refrigeration at 2°C-8°C
Dosage Ranges:
In combination with methotrexate, to reduce the signs and symptoms, inhibit the progression of structural damage, and improve physical function in patients with moderately to severely active rheumatoid arthritis: 3 mg/kg given as an intravenous infusion followed by additional doses at 2 and 6 weeks then every 8 weeks thereafter. Doses may be increased up to 10 mg/kg or given every 4 weeks.
To reduce the signs and symptoms and to induce and maintain clinical remission in patients with moderately to severely active Crohn’s disease who have had an inadequate response to conventional therapy AND to reduce the number of draining enterocutaneous and rectovaginal fistulas and to maintain fistula closure in patients with fistulizing Crohn’s disease: 5 mg/kg given as an intravenous infusion followed by additional doses at 2 and 6 weeks then 8 weeks thereafter. Doses may be increased up to 10 mg/kg. Patients who fail to respond within 14 weeks should discontinue therapy.
Administration and Stability: Infliximab may be used within 3 hours of reconstitution. Use 10 mL of Sterile Water for Injection, USP to dissolve the lyophilized powder. Vials should be gently swirled but not shaken. Allow the reconstitution to stand for 5 minutes. Further dilute to a total volume of 250 mL with 0.9% Sodium Chloride Injection, USP and with a final concentration of 0.4 mg/mL. The infusion should be administered over a period of not less than 2 hours using an infusion set with an in-line, sterile, non-pyrogenic, low protein-binding filter (pore size of 1.2 µm or less).
Pharmacology/Pharmacokinetics: Infliximab competitively blocks the binding of human tumor necrosis factor alpha (TNFa) to its receptors. Infliximab does not neutralize TNFb (lymphotoxin a), a related cytokine that utilizes the same receptors as TNFa. Elevated concentrations of TNFa have been found in the joints of rheumatoid arthritis patients and in the stools of Crohn’s disease patients. The biological activities of TNFa include induction of pro-inflammatory cytokines such as interleukins (IL) 1 and 6, enhancement of leukocyte migration by increasing endothelial layer permeability and expression of adhesion molecules by endothelial cells and leukocytes, activation of neutrophil and eosinophil functional activity, induction of acute phase reactants and other liver proteins, as well as tissue degrading enzymes produced by synoviocytes and/or chondrocytes. The terminal half-life of infliximab is around 9 days.
Drug and Lab Interactions: Concurrent administration of etanercept (another TNFa-blocking agent) and anakinra (an interleukin-1 antagonist) has been associated with an increased risk of serious infection. TNFa-blocking agents, including infliximab, may result in similar toxicities when used with anakinra.
Contraindications/Precautions: Contraindicated in patients with known hypersensitivity to any murine proteins or other components of this product. Doses of over 5 mg/kg are contraindicated in patients with moderate-to-severe heart failure. Serious infections, some fatal, have occurred in patients receiving TNF-blocking agents. Patients should be evaluated for latent tuberculosis infection with a tuberculin skin test. Do not use in patients with a clinically important, active infection. Use with caution in patients with a chronic infection or a history of recurrent infection. Discontinue use if a patient develops a serious infection. Use with caution in patients who have resided in regions where histoplasmosis or coccidioidomycosis is endemic. Cases of leukopenia, thrombocytopenia, and pancytopenia, some with fatal outcome, have been reported in patients receiving infliximab. Patients should seek immediate medical attention if they develop signs and symptoms of blood dyscrasias (e.g., persistent fever) while on infliximab. Use with caution in patients with preexisting central nervous system demyelinating or seizure disorders. Do not give live vaccines to patients using infliximab. No recommended in nursing mothers. Pregnancy Category B.
Monitoring Parameters: Vital signs, LFTs, s/s of infection, annual PPD
Adverse Effects: Acute infusion reactions, including fever, chills, chest pain, hypotension, dyspnea, hypertension, pruritus, and urticaria may occur with use. Infliximab increases the risk of developing infections, most commonly respiratory and urinary tract infections. Rare cases of lupus-like syndrome may occur. Less severe adverse effects include abdominal pain, dyspepsia, coughing, bronchitis, rhinitis, rash, pruritus, and headache. Crohn’s disease patients who were retreated with infliximab after a 2–4-year absence had a higher incidence of myalgia, arthralgia, pruritus, facial, hand, or lip edema, dysphagia, urticaria, sore throat, and headache.
Common Clinical Applications: Infliximab is used in treatment of rheumatoid arthritis, Crohn’s disease, ankylosing spondylitis, and treatment of and maintenance of ulcerative colitis that has not responded to conventional therapy.
2 notes
·
View notes
Text
subleq.fth
only forth definitions hex
system +order
variable meta.1
variable target.1
variable assembler.1
variable target.only.1
meta.1 +order definitions
2 constant =cell
4000 constant size
8000 constant =end
40 constant =stksz
100 constant =buf
0008 constant =bksp
000A constant =lf
000D constant =cr
007F constant =del
create tflash size cells here over erase allot
variable tdp
variable tep
variable tlast
size =cell - tep !
0 tlast !
: :m meta.1 +order definitions : ;
: ;m postpone ; ; immediate
:m there tdp @ ;m
:m tc! tflash + c! ;m
:m tc@ tflash + c@ ;m
:m t! over $FF and over tc! swap 8 rshift swap 1+ tc! ;m
:m t@ dup tc@ swap 1+ tc@ 8 lshift or ;m
:m taligned dup 1 and + ;m
:m talign there 1 and tdp +! ;m
:m tc, there tc! 1 tdp +! ;m
:m t, there t! 2 tdp +! ;m
:m $literal
talign [char] " word count dup tc, for aft count tc, then next drop talign ;m
:m tallot tdp +! ;m
:m parse-word bl word ?nul count ;m
:m thead
talign
there tlast @ t, tlast !
parse-word talign dup tc, for aft count tc, then next drop talign ;m
:m #dec dup >r abs 0 <# $A hold #s r> sign #> type ;m ( n -- print number )
:m mdump aligned begin ?dup while swap dup @ #dec cell+ swap cell - repeat drop ;m
:m save-target parse-word drop decimal tflash there mdump ;m
:m .end only forth definitions decimal ;m
:m setlast tlast ! ;m
:m atlast tlast @ ;m
:m tvar
get-current >r meta.1 set-current create r> set-current there , t, does> @ ;m
:m label:
get-current >r meta.1 set-current create r> set-current there , does> @ ;m
:m tdown =cell negate and ;m
:m tnfa =cell + ;m ( pwd -- nfa : move to name field address )
:m tcfa tnfa dup c@ $1F and + =cell + tdown ;m ( pwd -- cfa )
:m compile-only tlast @ tnfa t@ $20 or tlast @ tnfa t! ;m ( -- )
:m immediate tlast @ tnfa t@ $40 or tlast @ tnfa t! ;m ( -- )
:m half dup 1 and abort" unaligned" 2/ ;m
:m double 2* ;m
:m (') bl word find ?found cfa ;m
:m t' (') >body @ ;m
:m to' target.only.1 +order (') >body @ target.only.1 -order ;m
:m tcksum taligned dup $C0DE - $FFFF and >r
begin ?dup while swap dup t@ r> + $FFFF and >r =cell + swap =cell - repeat
drop r> ;m
:m mkck dup there swap - tcksum ;m
system -order
\ ---------------------------------- Forth VM --------------------------------
:m Z 0 t, ;m \ Address 0 must contain 0
:m NADDR there 2/ 1+ t, ;m
:m HALT 0 t, 0 t, -1 t, ;m
:m JMP 2/ Z Z t, ;m
:m ADD swap 2/ t, Z NADDR Z 2/ t, NADDR Z Z NADDR ;m
:m SUB swap 2/ t, 2/ t, NADDR ;m
:m NOP Z Z NADDR ;m
:m ZERO dup 2/ t, 2/ t, NADDR ;m
:m PUT 2/ t, -1 t, NADDR ;m
:m MOV 2/ >r r@ dup t, t, NADDR 2/ t, Z NADDR r> Z t, NADDR Z Z NADDR ;m
:m iLOAD there 2/ 3 4 * 3 + + 2* MOV 0 swap MOV ;m
:m iJMP there 2/ E + 2* MOV NOP ;m
:m iSTORE ( addr w -- )
swap >r there 2/ 24 + 2dup 2* MOV 2dup 1+ 2* MOV 7 + 2* MOV r> 0 MOV ;m
assembler.1 +order definitions
: begin talign there ;
: again JMP ;
: mark there 0 t, ;
: if ( NB. "if" does not work for 8000 )
2/ dup t, Z there 2/ 4 + dup t, Z Z 6 + t, Z Z NADDR Z t, mark ;
: until 2/ dup t, Z there 2/ 4 + dup t, Z Z 6 + t, Z Z NADDR Z t, 2/ t, ;
: +if Z 2/ t, mark ;
: -if 2/ t, Z there 2/ 4 + t, Z Z there 2/ 4 + t, Z Z mark ;
: then begin 2/ swap t! ;
: while if swap ;
: repeat JMP then ;
assembler.1 -order
meta.1 +order definitions
0 t, 0 t, \ both locations must be zero
label: entry \ used to set entry point in next cell
-1 t, \ system entry point
B tvar {options} \ bit #1=echo off, #2 = checksum on, #4=info, #8=die on EOF
0 tvar h \ dictionary pointer
0 tvar primitive \ any address lower than this one must be a primitive
0 tvar check \ used for system checksum
8000 tvar hbit \ must contain 8000
-2 tvar ntwo \ must contain -2
-1 tvar neg1 \ must contain -1
1 tvar one \ must contain 1
2 tvar two \ must contain 1
$10 tvar bwidth \ must contain 16
0 tvar INVREG \ temporary register used for inversion only
0 tvar {cold} \ entry point of virtual machine program, set later on
0 tvar {key} \ execution vector for key?
0 tvar {emit} \ execution vector for emit
0 tvar {literal} \ execution vector for literal
0 tvar {ok} \ execution vector for .ok
0 tvar {echo} \ execution vector for echo
0 tvar ip \ instruction pointer
0 tvar w \ working pointer
0 tvar x \ working pointer
0 tvar t \ temporary register for Virtual Machine
0 tvar bl1 \ bitwise extra register
0 tvar bl2 \ bitwise extra register
0 tvar bt \ bitwise extra register
0 tvar tos \ top of stack
0 tvar {state} \ compiler state
0 tvar {hld} \ hold space pointer
$A tvar {base} \ input/output radix
-1 tvar {dpl} \ number of places after fraction
0 tvar {in} \ position in query string
0 tvar {handler} \ throw/catch handler
0 tvar {last} \ last defined word
0 tvar #tib \ terminal input buffer
0 tvar {context} $E tallot \ vocabulary context
0 tvar {current} \ vocabulary which new definitions are added to
0 tvar {forth-wordlist} \ forth word list (main vocabulary)
0 tvar {root-voc} \ absolute minimum vocabulary
0 tvar {system} \ system functions vocabulary
=end dup tvar {sp0} tvar {sp} \ grows downwards
=end =stksz 4 * - dup tvar {rp0} tvar {rp} \ grows upwards
=end =stksz 4 * - =buf - constant TERMBUF \ pad buffer space
TERMBUF =buf + constant =tbufend
:m INC 2/ neg1 2/ t, t, NADDR ;m ( b -- )
:m DEC 2/ one 2/ t, t, NADDR ;m ( b -- )
:m INV ( b -- : invert NB. b - a = b + ~a + 1 )
INVREG ZERO dup INVREG SUB dup INVREG swap MOV DEC ;m
:m ++sp {sp} DEC ;m
:m --sp {sp} INC ;m
:m --rp {rp} DEC ;m
:m ++rp {rp} INC ;m
:m a-optim >r there =cell - r> 2/ t! ;m
assembler.1 +order
label: start
start 2/ entry t!
{sp0} {sp} MOV
{rp0} {rp} MOV
{cold} ip MOV
( fall-through )
label: vm ( Forth Inner Interpreter )
ip w MOV
ip INC
t w iLOAD
t w MOV
primitive t SUB
t -if w iJMP then ( jump straight to VM functions )
++rp
ip {rp} iSTORE
w ip MOV vm a-optim
vm JMP
assembler.1 -order
:m header >in @ thead >in ! ;m
:m :ht ( "name" -- : forth routine, no header )
get-current >r target.1 set-current create
r> set-current $BABE talign there ,
does> @ 2/ t, ;m
:m :t header :ht ;m ( "name" -- : forth routine )
:m :to ( "name" -- : forth, target only routine )
header
get-current >r target.only.1 set-current create r> set-current
$BABE talign there ,
does> @ 2/ t, ;m
:m :a ( "name" -- : assembly routine, no header )
D00D
target.1 +order definitions
create talign there , assembler.1 +order does> @ 2/ t, ;m
:m (a); D00D <> if abort" unstructured" then assembler.1 -order ;m
:m ;a (a); vm a-optim vm JMP ;m
:m postpone target.only.1 +order t' target.only.1 -order 2/ t, ;m
\ NB. There are some bugs with the comparison operators "op<" and "op>" when
\ they deal with extreme values like "$8000 1 <", "$8002 1 <" words fine.
:a bye HALT ;a
:a 1- tos DEC ;a
:a 1+ tos INC ;a
:a invert tos INV ;a
:a [@] tos tos iLOAD ;a
:a [!] w {sp} iLOAD w tos iSTORE --sp tos {sp} iLOAD --sp ;a
:a opEmit tos PUT tos {sp} iLOAD --sp ;a
:a opPush ++sp tos {sp} iSTORE tos ip iLOAD ip INC ;a
:a opSwap tos w MOV tos {sp} iLOAD w {sp} iSTORE ;a
:a opDup ++sp tos {sp} iSTORE ;a
:a opOver w {sp} iLOAD ++sp tos {sp} iSTORE w tos MOV ;a
:a opDrop tos {sp} iLOAD --sp ;a
:a opToR ++rp tos {rp} iSTORE tos {sp} iLOAD --sp ;a
:a opFromR ++sp tos {sp} iSTORE tos {rp} iLOAD --rp ;a
:a opMul w {sp} iLOAD t ZERO
begin w while tos t ADD w DEC repeat t tos MOV --sp ;a
:a opExit ip {rp} iLOAD --rp ;a
:a - w {sp} iLOAD tos w SUB w tos MOV --sp ;a
:a + w {sp} iLOAD w tos ADD --sp ;a
:a r@ ++sp tos {sp} iSTORE tos {rp} iLOAD ;a
:a rdrop --rp ;a
:a sp@ ++sp tos {sp} iSTORE {sp} tos MOV tos INC ;a
:a sp! tos {sp} MOV ;a
:a rp@ ++sp tos {sp} iSTORE {rp} tos MOV ;a
:a rp! tos {rp} MOV tos {sp} iLOAD --sp ;a
:a opNext w {rp} iLOAD
w if w DEC w {rp} iSTORE t ip iLOAD t ip MOV vm JMP then ip INC --rp ;a
:a lsb
tos tos ADD tos tos ADD tos tos ADD tos tos ADD
tos tos ADD tos tos ADD tos tos ADD tos tos ADD
tos tos ADD tos tos ADD tos tos ADD tos tos ADD
tos tos ADD tos tos ADD
tos w MOV 0 tos MOV w if neg1 tos MOV then ;a
:a opJump ip ip iLOAD ;a
:a opJumpZ
tos w MOV 0 t MOV w if neg1 t MOV then w DEC w +if neg1 t MOV then
tos {sp} iLOAD --sp t if ip INC vm JMP then w ip iLOAD w ip MOV ;a
:a op0> tos w MOV 0 tos MOV w +if neg1 tos MOV then ;a
:a op0=
tos w MOV neg1 tos MOV w if 0 tos MOV then w DEC w +if 0 tos MOV then ;a
:a op0<
tos w MOV 0 tos MOV w -if neg1 tos MOV then w INC w -if neg1 tos MOV then ;a
:a op< w {sp} iLOAD --sp tos w SUB 0 tos MOV w -if neg1 tos MOV then ;a
:a op> w {sp} iLOAD --sp tos w SUB 0 tos MOV w +if neg1 tos MOV then ;a
:a op2* tos tos ADD ;a
:a op2/
bwidth w MOV
x ZERO
begin w DEC w while
x x ADD
tos bt MOV 0 bl1 MOV bt -if neg1 bl1 MOV then bt INC bt -if neg1 bl1 MOV then
bl1 if x INC then
tos tos ADD
repeat
x tos MOV ;a
:a rshift
bwidth w MOV
tos w SUB
tos {sp} iLOAD --sp
x ZERO
begin w while
x x ADD
tos bt MOV 0 bl1 MOV bt -if neg1 bl1 MOV then bt INC bt -if neg1 bl1 MOV then
bl1 if x INC then
tos tos ADD
w DEC
repeat
x tos MOV ;a
:a opOr
bwidth w MOV
x ZERO
t {sp} iLOAD
--sp
begin w while
x x ADD
tos bt MOV 0 bl1 MOV bt -if neg1 bl1 MOV then bt INC bt -if neg1 bl1 MOV then
t bt MOV 0 bl2 MOV bt -if neg1 bl2 MOV then bt INC bt -if neg1 bl2 MOV then
bl1 bl2 ADD bl2 if x INC then
t t ADD
tos tos ADD
w DEC
repeat
x tos MOV ;a
:a opXor
bwidth w MOV
x ZERO
t {sp} iLOAD
--sp
begin w while
x x ADD
tos bt MOV 0 bl1 MOV bt -if neg1 bl1 MOV then bt INC bt -if neg1 bl1 MOV then
t bt MOV 0 bl2 MOV bt -if neg1 bl2 MOV then bt INC bt -if neg1 bl2 MOV then
bl1 bl2 ADD bl2 INC one bl1 MOV bl2 if 0 bl1 MOV then bl1 x ADD
t t ADD
tos tos ADD
w DEC
repeat
x tos MOV ;a
:a opAnd
bwidth w MOV
x ZERO
t {sp} iLOAD
--sp
begin w while
x x ADD
tos bt MOV 0 bl1 MOV bt -if neg1 bl1 MOV then bt INC bt -if neg1 bl1 MOV then
t bt MOV 0 bl2 MOV bt -if neg1 bl2 MOV then bt INC bt -if neg1 bl2 MOV then
bl1 bl2 ADD two bl2 ADD one bl1 MOV bl2 if 0 bl1 MOV then bl1 x ADD
t t ADD
tos tos ADD
w DEC
repeat
x tos MOV ;a
there 2/ primitive t!
:m ;t $BABE <>
if abort" unstructured" then talign opExit target.only.1 -order ;m
:m :s tlast @ {system} t@ tlast ! $F00D :t drop 0 ;m
:m :so tlast @ {system} t@ tlast ! $F00D :to drop 0 ;m
:m ;s drop $BABE ;t $F00D <> if abort" unstructured" then
tlast @ {system} t! tlast ! ;m
:m :r tlast @ {root-voc} t@ tlast ! $F00D :t drop 0 ;m
:m ;r drop $BABE ;t $F00D <> if abort" unstructured" then
tlast @ {root-voc} t! tlast ! ;m
:m lit opPush t, ;m
:m [char] char opPush t, ;m
:m char char opPush t, ;m
:m begin talign there ;m
:m until talign opJumpZ 2/ t, ;m
:m again talign opJump 2/ t, ;m
:m if opJumpZ there 0 t, ;m
:m mark opJump there 0 t, ;m
:m then there 2/ swap t! ;m
:m else mark swap then ;m
:m while if ;m
:m repeat swap again then ;m
:m aft drop mark begin swap ;m
:m next talign opNext 2/ t, ;m
:m for opToR begin ;m
:m =push [ t' opPush half ] literal ;m
:m =jump [ t' opJump half ] literal ;m
:m =jumpz [ t' opJumpZ half ] literal ;m
:m =unnest [ t' opExit half ] literal ;m
:m =>r [ t' opToR half ] literal ;m
:m =next [ t' opNext half ] literal ;m
:m dup opDup ;m
:m drop opDrop ;m
:m over opOver ;m
:m swap opSwap ;m
:m >r opToR ;m
:m r> opFromR ;m
:m 0> op0> ;m
:m 0= op0= ;m
:m 0< op0< ;m
:m < op< ;m
:m > op> ;m
:m or opOr ;m
:m xor opXor ;m
:m and opAnd ;m
:m exit opExit ;m
:m 2/ op2/ ;m
:ht #0 0 lit ;t
:ht #1 1 lit ;t
:ht #-1 FFFF lit ;t
:to 1+ 1+ ;t
:to 1- 1- ;t
:to + + ;t
:to - - ;t
:to invert invert ;t
:to bye bye ;t
:to dup dup ;t
:to drop opDrop ;t
:to over opOver ;t
:to swap opSwap ;t
:to rshift rshift ;t
:so [@] [@] ;s
:so [!] [!] ;s
:so lsb lsb ;s
:to sp@ sp@ ;t
:to sp! sp! ;t
:to 0> op0> ;t
:to 0= op0= ;t
:to 0< op0< ;t
:to < op< ;t
:to > op> ;t
:to 2/ op2/ ;t
:to or opOr ;t
:to xor opXor ;t
:to and opAnd ;t
:to * opMul ;t
:t nop ;t
:t <ok> {ok} lit ;t
:s <emit> {emit} lit ;s
:s <key> {key} lit ;s
:s <literal> {literal} lit ;s
:s <cold> {cold} lit ;s
:s <echo> {echo} lit ;s
:t current {current} lit ;t
:t root-voc {root-voc} lit ;t
:t #vocs 8 lit ;t
:t context {context} lit ;t
:t here h half lit [@] ;t
:t base {base} lit ;t
:t dpl {dpl} lit ;t
:t hld {hld} lit ;t
:t bl 20 lit ;t
:t >in {in} lit ;t
:t hex $10 lit {base} half lit [!] ;t
:t decimal $A lit {base} half lit [!] ;t
:t source TERMBUF lit #tib half lit [@] ;t
:s many #0 {in} half lit [!] ;s
:t state {state} lit ;t
:t ] #-1 {state} half lit [!] ;t
:t [ #0 {state} half lit [!] ;t immediate
:t nip swap drop ;t
:t tuck swap over ;t
:t ?dup dup if dup then ;t
:t rot >r swap r> swap ;t
:t -rot rot rot ;t
:t 2drop drop drop ;t
:t 2dup over over ;t
:t 0<= 0> 0= ;t
:t 0<> 0= 0= ;t
:t = - 0= ;t
:t <> = 0= ;t
:t >= < 0= ;t
:t <= > 0= ;t
:t 0>= 0< 0= ;t
:t negate 1- invert ;t
:t s>d dup 0< ;t
:t abs s>d if negate then ;t
:t 2* op2* ;t
:t cell 2 lit ;t
:t cell+ cell + ;t
:t cells op2* ;t
:t u< 2dup 0< 0= swap 0< 0= <> >r < r> <> ;t
:t u> swap u< ;t
:t u>= u< 0= ;t
:t u<= u> 0= ;t
:t execute 2/ >r ;t
:t key? #-1 [@] negate s>d if
{options} half lit [@] 8 lit and if bye then drop #0 exit then #-1 ;t
:t key begin {key} half lit [@] execute until ;t
:t emit {emit} half lit [@] execute ;t
:t cr =cr lit emit =lf lit emit ;t
:t @ 2/ [@] ;t
:t ! 2/ [!] ;t
:t get-current current @ ;t
:t set-current current ! ;t
:s last get-current @ ;s
:t pick sp@ + [@] ;t
:t +! 2/ tuck [@] + swap [!] ;t
:t lshift begin ?dup while 1- swap 2* swap repeat ;t
:t c@ dup @ swap lsb if 8 lit rshift else $FF lit and then ;t
:t c! swap $FF lit and dup 8 lit lshift or swap
tuck dup @ swap lsb 0= $FF lit xor
>r over xor r> and xor swap ! ;t
:t max 2dup < if nip else drop then ;t
:t min 2dup > if nip else drop then ;t
:t count dup 1+ swap c@ ;t
:s logical 0<> if #1 else #0 then ;s
:t aligned dup lsb logical + ;t
:t align here aligned h half lit [!] ;t
:t +string #1 over min rot over + rot rot - ;t
:t type begin dup while swap count emit swap 1- repeat 2drop ;t
:t cmove for aft >r dup c@ r@ c! 1+ r> 1+ then next 2drop ;t ( b1 b2 u -- )
:t fill swap for swap aft 2dup c! 1+ then next 2drop ;t ( b u c -- )
:t erase #0 fill ;t ( NB. blank is bl fill )
:s do$ r> r> 2* dup count + aligned 2/ >r swap >r ;s ( -- a : )
:s ($) do$ ;s ( -- a : do string NB. )
:s .$ do$ count type ;s ( -- : print string, next cells contain string )
:m ." .$ $literal ;m
:m $" ($) $literal ;m
:t space bl emit ;t
:t catch ( xt -- exception# | 0 \ return addr on stack )
sp@ >r ( xt ) \ save data stack pointer
{handler} half lit [@] >r ( xt ) \ and previous handler
rp@ {handler} half lit [!] ( xt ) \ set current handler
execute ( ) \ execute returns if no throw
r> {handler} half lit [!] ( ) \ restore previous handler
rdrop ( ) \ discard saved stack ptr
#0 ;t ( 0 ) \ normal completion
:t throw ( ??? exception# -- ??? exception# )
?dup if ( exc# ) \ 0 throw is no-op
{handler} half lit [@] rp! ( exc# ) \ restore prev return stack
r> {handler} half lit [!] ( exc# ) \ restore prev handler
r> swap >r ( saved-sp ) \ exc# on return stack
sp! drop r> �� ( exc# ) \ restore stack
then ;t
:t abort #-1 throw ;t
:s (abort) do$ swap if count type abort then drop ;s
:t um+
2dup + >r r@ #0 >= >r 2dup and 0< r> or >r or 0< r> and invert 1+ r> swap ;t
:t dnegate invert >r invert #1 um+ r> + ;t ( d -- d )
:t d+ >r swap >r um+ r> + r> + ;t ( d d -- d )
:t um* ( u u -- ud : double cell width multiply )
#0 swap ( u1 0 u2 ) $F lit
for
dup um+ >r >r dup um+ r> + r>
if >r over um+ r> + then
next rot drop ;t
:t um/mod ( ud u -- ur uq : unsigned double cell width divide/modulo )
?dup 0= if -A lit throw then
2dup u<
if negate $F lit
for >r dup um+ >r >r dup um+ r> + dup
r> r@ swap >r um+ r> ( or -> ) 0<> swap 0<> +
if >r drop 1+ r> else drop then r>
next
drop swap exit
then 2drop drop #-1 dup ;t
:t m/mod ( d n -- r q : floored division )
s>d dup >r
if
negate >r dnegate r>
then
>r s>d if r@ + then r> um/mod r>
if swap negate swap exit then ;t
:t /mod over 0< swap m/mod ;t
:t mod /mod drop ;t
:t / /mod nip ;t
:s depth {sp0} half lit [@] sp@ - 1- ;s
:s (emit) opEmit ;s
:t echo {echo} half lit [@] execute ;t
:s tap dup echo over c! 1+ ;s ( bot eot cur c -- bot eot cur )
:s ktap ( bot eot cur c -- bot eot cur )
dup dup =cr lit <> >r =lf lit <> r> and if ( Not End of Line? )
dup =bksp lit <> >r =del lit <> r> and if ( Not Delete Char? )
bl tap
exit
then
>r over r@ < dup if
=bksp lit dup echo bl echo echo
then
r> +
exit
then drop nip dup ;s
:t accept ( b u -- b u : read in a line of user input )
over + over begin
2dup <>
while
key dup bl - $5F lit u< if tap else ktap then
repeat drop over - ;t
:t query TERMBUF lit =buf lit accept #tib lit ! drop #0 >in ! ;t
:s ?depth depth > if -4 lit throw then ;s ( u -- : check stack depth )
:t -trailing for aft bl over r@ + c@ < if r> 1+ exit then then next #0 ;t
:s look ( b u c xt -- b u : skip until *xt* test succeeds )
swap >r rot rot
begin
dup
while
over c@ r@ - r@ bl = 4 lit pick execute
if rdrop rot drop exit then
+string
repeat rdrop rot drop ;s
:s unmatch if 0> exit then 0<> ;s ( c1 c2 -- t )
:s match unmatch invert ;s ( c1 c2 -- t )
:t parse ( c -- b u ; <string> )
>r source drop >in @ + #tib lit @ >in @ - r@
>r over r> swap >r >r
r@ t' unmatch lit look 2dup
r> t' match lit look swap r> - >r - r> 1+ ( b u c -- b u delta )
>in +!
r> bl = if -trailing then #0 max ;t
:t spaces begin dup 0> while space 1- repeat drop ;t ( +n -- )
:t hold #-1 hld +! hld @ c! ;t ( c -- : save a character in hold space )
:t #> 2drop hld @ =tbufend lit over - ;t ( u -- b u )
:s extract dup >r um/mod r> swap >r um/mod r> rot ;s ( ud ud -- ud u )
:s digit 9 lit over < 7 lit and + [char] 0 + ;s ( u -- c )
:t # 2 lit ?depth #0 base @ extract digit hold ;t ( d -- d )
:t #s begin # 2dup ( d0= -> ) or 0= until ;t ( d -- 0 )
:t <# =tbufend lit hld ! ;t ( -- )
:t sign 0< if [char] - hold then ;t ( n -- )
:t u.r >r #0 <# #s #> r> over - spaces type ;t
:t u. #0 <# #s #> space type ;t
:t . dup >r abs #0 <# #s r> sign #> space type ;t ( n -- print number )
:t >number ( ud b u -- ud b u : convert string to number )
begin
2dup >r >r drop c@ base @ ( get next character )
( digit? -> ) >r [char] 0 - 9 lit over <
if 7 lit - dup $A lit < or then dup r> u< ( c base -- u f )
0= if ( d char )
drop ( d char -- d )
r> r> ( restore string )
exit ( ..exit )
then ( d char )
swap base @ um* drop rot base @ um* d+ ( accumulate digit )
r> r> ( restore string )
+string dup 0= ( advance string and test for end )
until ;t
:t number? ( a u -- d -1 | a u 0 : string to a number [easier to use] )
#-1 dpl !
base @ >r
over c@ [char] - = dup >r if +string then
over c@ [char] $ = if hex +string then
>r >r #0 dup r> r>
begin
>number dup
while over c@ [char] . <>
if rot drop rot r> 2drop #0 r> base ! exit then
1- dpl ! 1+ dpl @
repeat
2drop r> if dnegate then r> base ! #-1 ;t
:t compare ( a1 u1 a2 u2 -- n : string equality )
rot
over - ?dup if >r 2drop r> nip exit then
for ( a1 a2 )
aft
count rot count rot - ?dup
if rdrop nip nip exit then
then
next 2drop #0 ;t
:t .s depth for aft r@ pick . then next ;t
:t nfa cell+ ;t ( pwd -- nfa : move word pointer to name field )
:t cfa nfa dup c@ $1F lit and + cell+ cell negate and ;t ( pwd -- cfa )
:t allot aligned h lit +! ;t
:t , align here ! cell allot ;t
:s (search-wordlist) ( a wid -- PWD PWD 1|PWD PWD -1|0 a 0: find word in WID )
swap >r dup
begin
dup
while
dup nfa count $9F lit ( $1F:word-length + $80:hidden )
and r@ count compare 0=
if ( found! )
rdrop
dup ( immediate? -> ) nfa $40 lit swap @ and 0<>
#1 or negate exit
then
nip dup @
repeat
rdrop 2drop #0 ;s
:s (find) ( a -- pwd pwd 1 | pwd pwd -1 | 0 a 0 : find a word dictionary )
>r
context
begin
dup @
while
dup @ @ r@ swap (search-wordlist) ?dup
if
>r rot drop r> rdrop exit
then
cell+
repeat drop #0 r> #0 ;s
:t search-wordlist (search-wordlist) rot drop ;t ( a wid -- PWD 1|PWD -1|a 0 )
:t find ( a -- pwd 1 | pwd -1 | a 0 : find a word in the dictionary )
(find) rot drop ;t
:s (literal) state @ if =push lit , , then ;s
:t literal <literal> @ execute ;t immediate ( u -- )
:t compile, 2/ align , ;t ( xt -- )
:s ?found
if exit then space count type [char] ? emit cr -D lit throw ;s ( u f -- )
:t interpret ( b -- )
find ?dup if
state @
if
0> if cfa execute exit then \ <- immediate word are executed
cfa compile, exit \ <- compiling word are...compiled.
then
drop
dup nfa c@ 20 lit and if -E lit throw then ( <- ?compile )
cfa execute exit \ <- if its not, execute it, then exit *interpreter*
then
\ not a word
dup >r count number? if rdrop \ it is a number!
dpl @ 0< if \ <- dpl will be -1 if it is a single cell number
drop \ drop high cell from 'number?' for single cell output
else \ <- dpl is not -1, it is a double cell number
state @ if swap then
postpone literal \ literal is executed twice if it's a double
then
postpone literal exit
then
r> #0 ?found ;t \ Could vector ?found here, to handle arbitrary words
:s .id nfa count $1F lit and type space ;s ( pwd -- : print out a word )
:t get-order ( -- widn ... wid1 n : get the current search order )
context
( find empty cell -> ) #0 >r begin dup @ r@ xor while cell+ repeat rdrop
dup cell - swap
context - 2/ dup >r 1- s>d if -50 lit throw then
for aft dup @ swap cell - then next @ r> ;t
:r set-order ( widn ... wid1 n -- : set the current search order )
dup #-1 = if drop root-voc #1 set-order exit then ( NB. Recursion! )
dup #vocs > if -49 lit throw then
context swap for aft tuck ! cell+ then next #0 swap ! ;r
:r forth-wordlist {forth-wordlist} lit ;r ( -- wid )
:r system {system} lit ;r ( -- wid )
:r forth root-voc forth-wordlist 2 lit set-order ;r ( -- )
:r only #-1 set-order ;r ( -- )
:r words
get-order begin ?dup while swap dup cr u. ." : " @
begin ?dup while dup nfa c@ $80 lit and 0= if dup .id then @ repeat cr
1- repeat ;r
:t definitions context @ set-current ;t
:t (order) ( w wid*n n -- wid*n w n )
dup if
1- swap >r (order) over r@ xor
if
1+ r> -rot exit
then rdrop
then ;t
:t -order get-order (order) nip set-order ;t
:t +order dup >r -order get-order r> swap 1+ set-order ;t
:t word parse here dup >r 2dup ! 1+ swap cmove r> ;t ( c -- b )
:s ?unique ( a -- a : print a message if a word definition is not unique )
dup get-current (search-wordlist) 0= if exit then space
2drop {last} lit @ .id ." redefined" cr ;s
:s ?nul dup c@ if exit then -10 lit throw ;s
:to char bl word ?nul count drop c@ ;t
:to [char] postpone char =push lit , , ;t immediate
:to ; $BABE lit <> if -16 lit throw then =unnest lit , postpone [
?dup if get-current ! exit then ;t immediate compile-only ( -- wid )
:to : align here dup {last} lit ! ( "name", -- colon-sys )
last , bl word ?nul ?unique count + h lit ! align $BABE lit postpone ] ;t
:to :noname here $BABE lit ] ;t
:to begin align here ;t immediate compile-only
:to until =jumpz lit , 2/ , ;t immediate compile-only
:to again =jump lit , 2/ , ;t immediate compile-only
:to if =jumpz lit , here #0 , ;t immediate compile-only
:to then here 2/ swap ! ;t immediate compile-only
:to while postpone if ;t immediate compile-only
:to repeat swap postpone again postpone then ;t immediate compile-only
:to else =jump lit , here #0 , swap postpone then ;t immediate compile-only
:to for =>r lit , here ;t immediate compile-only
:to aft drop =jump lit , here #0 , align here swap ;t immediate compile-only
:to next =next lit , 2/ , ;t immediate compile-only
:to ' bl word find ?found cfa literal ;t immediate
:t compile r> dup [@] , 1+ >r ;t compile-only
:t recurse {last} lit @ cfa compile, ;t immediate compile-only
:s toggle tuck @ xor swap ! ;s
:s hide bl word find ?found nfa $80 lit swap toggle ;s
:s (var) r> 2* ;s compile-only
:s (const) r> [@] ;s compile-only
:s (marker) r> 2* dup @ h lit ! cell+ @ get-current ! ;s compile-only
:t create postpone : drop postpone [ compile (var) get-current ! ;t
:to variable create #0 , ;t
:to constant create cell negate allot compile (const) , ;t
:to marker last here create cell negate allot compile (marker) , , ;t
:t >body cell+ ;t ( a -- a )
:s (does) r> r> 2* swap >r ;s compile-only
:s (comp) r> {last} lit @ cfa ! ;s compile-only
:t does> compile (comp) compile (does) ;t immediate compile-only
:to rp! compile rp! ;t immediate compile-only
:to rp@ compile rp@ ;t immediate compile-only
:to >r compile opToR ;t immediate compile-only
:to r> compile opFromR ;t immediate compile-only
:to r@ compile r@ ;t immediate compile-only
:to rdrop compile rdrop ;t immediate compile-only
:to exit compile opExit ;t immediate compile-only
:to ." compile .$
[char] " word count + h half lit [!] align ;t immediate compile-only
:to $" compile ($)
[char] " word count + h half lit [!] align ;t immediate compile-only
:to abort" compile (abort)
[char] " word count + h half lit [!] align ;t immediate compile-only
:to ( [char] ) parse 2drop ;t immediate
:to .( [char] ) parse type ;t immediate
:to postpone bl word find ?found cfa compile, ;t immediate
:to ) ;t immediate
:to \ source drop @ {in} half lit [!] ;t immediate
:to immediate last nfa @ $40 lit or last nfa ! ;t
:to see bl word find ?found
cr begin dup @ =unnest lit <> while dup @ u. cell+ repeat @ u. ;t
:to dump aligned begin ?dup while swap dup @ . cell+ swap cell - repeat drop ;t
:s cksum aligned dup $C0DE lit - >r
begin ?dup while swap dup @ r> + >r cell+ swap cell - repeat drop r> ;s
:s ok state @ 0= if ." ok" cr then ;s
:t eval
begin bl word dup c@ while
interpret #1 ?depth
repeat drop {ok} half lit [@] execute ;t ( "word" -- )
:s info cr
." Project: eForth v1.5 " ( here . ) cr
." Author: Richard James Howe" cr
." Email: [email protected]" cr
." Repo: https://github.com/howerj/subleq" cr
." License: The Unlicense / Public Domain" cr ;s
:s ini only forth definitions decimal postpone [
#0 {in} half lit [!] #-1 {dpl} half lit [!] ;s ( -- )
:s opts
{options} half lit [@] lsb if to' drop lit {echo} half lit [!] then
{options} half lit [@] 4 lit and if info then
{options} half lit [@] 2 lit and if
primitive half lit [@] 2* dup here swap - cksum
check half lit [@] <> if ." cksum fail" bye then
{options} half lit [@] 2 lit xor {options} half lit [!]
then ;s
:t quit ( -- : interpreter loop, and more, does more than most QUITs )
ini
opts
begin
query t' eval lit catch
?dup if dup space . [char] ? emit cr #-1 = if bye then ini then
again ;t
:t cold {cold} half lit [@] execute ;t
\ ---------------------------------- Image Generation ------------------------
t' quit half {cold} t!
t' key? {key} t!
t' (emit) {echo} t!
t' (emit) {emit} t!
t' ok {ok} t!
t' (literal) {literal} t!
atlast {forth-wordlist} t!
{forth-wordlist} {current} t!
there h t!
primitive t@ double mkck check t!
atlast {last} t!
save-target subleq.dec
there .end
bye
0 notes