#RNA sequencing
Explore tagged Tumblr posts
Text

Exploring RNA Interference
Imagine a molecular switch within your cells, one that can selectively turn off the production of specific proteins. This isn't science fiction; it's the power of RNA interference (RNAi), a groundbreaking biological process that has revolutionized our understanding of gene expression and holds immense potential for medicine and beyond.
The discovery of RNAi, like many scientific breakthroughs, was serendipitous. In the 1990s, Andrew Fire and Craig Mello were studying gene expression in the humble roundworm, Caenorhabditis elegans (a tiny worm). While injecting worms with DNA to study a specific gene, they observed an unexpected silencing effect - not just in the injected cells, but throughout the organism. This puzzling phenomenon, initially named "co-suppression," was later recognized as a universal mechanism: RNAi.
Their groundbreaking work, awarded the Nobel Prize in 2006, sparked a scientific revolution. Researchers delved deeper, unveiling the intricate choreography of RNAi. Double-stranded RNA molecules, the key players, bind to a protein complex called RISC (RNA-induced silencing complex). RISC, equipped with an "Argonaut" enzyme, acts as a molecular matchmaker, pairing the incoming RNA with its target messenger RNA (mRNA) - the blueprint for protein production. This recognition triggers the cleavage of the target mRNA, effectively silencing the corresponding gene.
So, how exactly does RNAi silence genes? Imagine a bustling factory where DNA blueprints are used to build protein machines. RNAi acts like a tiny conductor, wielding double-stranded RNA molecules as batons. These batons bind to specific messenger RNA (mRNA) molecules, the blueprints for proteins. Now comes the clever part: with the mRNA "marked," special molecular machines chop it up, effectively preventing protein production. This targeted silencing allows scientists to turn down the volume of specific genes, observing the resulting effects and understanding their roles in health and disease.
The intricate dance of RNAi involves several key players:dsRNA: The conductor, a long molecule with two complementary strands. Dicer: The technician, an enzyme that chops dsRNA into small interfering RNAs (siRNAs), about 20-25 nucleotides long. RNA-induced silencing complex (RISC): The ensemble, containing Argonaute proteins and the siRNA. Target mRNA: The specific "instrument" to be silenced, carrying the genetic instructions for protein synthesis.
The siRNA within RISC identifies and binds to the complementary sequence on the target mRNA. This binding triggers either:Direct cleavage: Argonaute acts like a molecular scissors, severing the mRNA, preventing protein production. Translation inhibition: RISC recruits other proteins that block ribosomes from translating the mRNA into a protein.
From Labs to Life: The Diverse Applications of RNAi
The ability to silence genes with high specificity unlocks various applications across different fields:
Unlocking Gene Function: Researchers use RNAi to study gene function in various organisms, from model systems like fruit flies to complex human cells. Silencing specific genes reveals their roles in development, disease, and other biological processes.
Therapeutic Potential: RNAi holds immense promise for treating various diseases. siRNA-based drugs are being developed to target genes involved in cancer, viral infections, neurodegenerative diseases, and more. Several clinical trials are underway, showcasing the potential for personalized medicine.
Crop Improvement: In agriculture, RNAi offers sustainable solutions for pest control and crop development. Silencing genes in insects can create pest-resistant crops, while altering plant genes can improve yield, nutritional value, and stress tolerance.
Beyond the Obvious: RNAi applications extend beyond these core areas. It's being explored for gene therapy, stem cell research, and functional genomics, pushing the boundaries of scientific exploration.
Despite its exciting potential, RNAi raises ethical concerns. Off-target effects, unintended silencing of non-target genes, and potential environmental risks need careful consideration. Open and responsible research, coupled with public discourse, is crucial to ensure we harness this powerful tool for good.
RNAi, a testament to biological elegance, has revolutionized our understanding of gene regulation and holds immense potential for transforming various fields. As advancements continue, the future of RNAi seems bright, promising to silence not just genes, but also diseases, food insecurity, and limitations in scientific exploration. The symphony of life, once thought unchangeable, now echoes with the possibility of fine-tuning its notes, thanks to the power of RNA interference.
#science sculpt#life science#science#molecular biology#biology#biotechnology#dna#double helix#genetics#artists on tumblr#rna#rna sequencing#RNA interference#cell biology#cells#biomolecules#illustrates#scientific illustration#illustration#illustrative art#scientific research
12 notes
·
View notes
Text

Resident Upheaval
Using imaging mass cytometry and single-cell RNA sequencing to analyse the cellular composition and topology of the tissue changes with ulcerative colitis – inflammatory macrophages replace resident macrophages
Read the original article here
Image from work by Juan Du, Junlei Zhang, Lin Wang and Xun Wang, and colleagues
Zhejiang University School of Medicine, Hangzhou, China
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in Nature Communications, June 2023
You can also follow BPoD on Instagram, Twitter and Facebook
5 notes
·
View notes
Text
Unlocking Insights: A Beginner’s Guide to Gene Expression Profiling and Pathway Enrichment
In the field of genomics, understanding the behavior of genes is essential to unlocking the mysteries of biology and disease. Two important techniques, gene expression profiling and pathway enrichment analysis, help researchers unravel the complexity of genetic regulation, allowing for a deeper understanding of biological processes, disease mechanisms, and therapeutic targets. This article provides a beginner’s guide to gene expression profiling and pathway enrichment analysis, explaining their significance, methods, and applications.
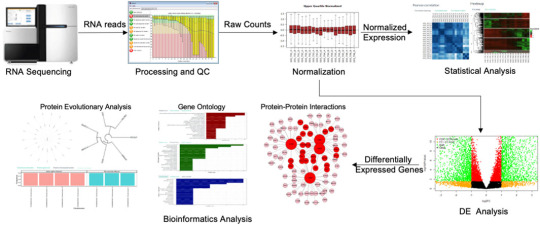
To explore more about applications of Gene Expression Profiling and Pathway Enrichment Analysis, you can visit Gene-omics.
If you want to learn about the genomics from the beginning, you can enroll for the Genome Analysis Bootcamp Demo Session here- https://edgeneomics.com/genome-analysis-bootcamp/
0 notes
Text
Central Dogma of Life
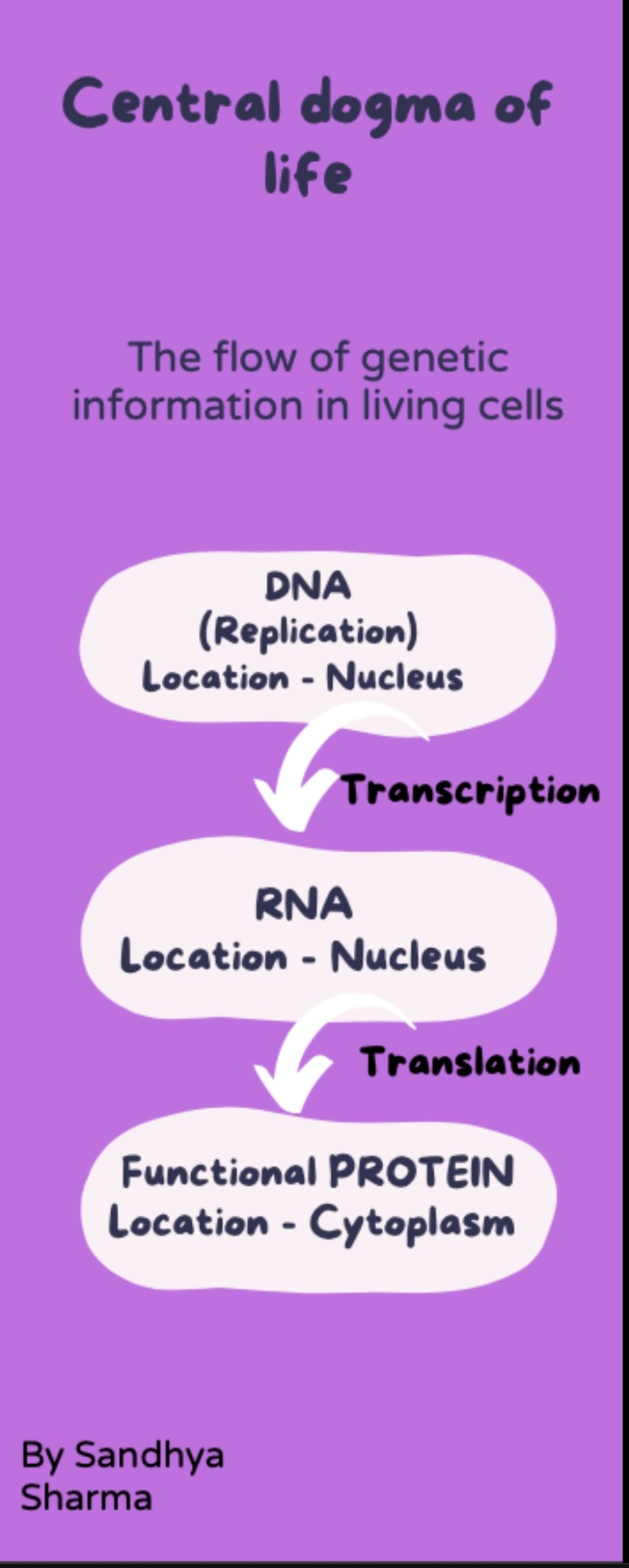
#dna#chemistry#science#dna activation#biotechnology#biochemistry#biology#rna#rna sequencing#replication#protein#infographic#useful#resources#info post#information#helpful#science communication#science class#life science#genetics#cells at work
1 note
·
View note
Text

Me when my teacher supervisor promised to warn me when the course inscription open and she didn't.
Now I need to find somebody to teach me rna-seq. Send help please oh my god is for my thesis
1 note
·
View note
Text
Dr. McCullough demands the EU, US pull out of World Health Organization over COVID response - LifeSite
#covid19#covid#sars cov 2#rna sequencing#rna therapeutics#vaccine#great reset#agenda 2030#World Economic Forum#Eurasian Economic Forum#Rothchild
1 note
·
View note
Text
Scientists have extracted RNA from an extinct species for the first time!!
They extracted it from a mummified Thylacine (Tasmanian Tiger) that was stored in Stockholm.
Just 2 months after the Thylacine was granted a protection status, the last of its kind passed from exposure in 1936 at the Beaumaris Zoo in Hobart.
It was the world's largest carnivorous marsupial, estimated to stand as tall as 51 inches and weighing around 66lb.
I'm very excited for the potential of bringing this species back - Jurassic Park style. We hunted this creature to extinction and now we may be able to fix that.
#thylacine#tasmanian tiger#science#rna sequencing#rna extraction#extinct animals#extinct species#extinction
1 note
·
View note
Text
Happy to share, ARTEMIS is now published at NAR!
https://doi.org/10.1093/nar/gkae758
ARTEMIS is a new tool for RNA/DNA 3D structure superposition and structure-based sequence alignment. Our benchmarks show that it outperforms the existing tools for both sequentially-ordered and topology-independent alignment.
ARTEMIS allowed us to identify an intriguing structural similarity between Lysine and M-box riboswitches (see the figure) and to describe the minor-groove/minor-groove helical packing motif. ARTEMIS is the first tool able to report several alternative superpositions, which makes it suitable for structural motif identification tasks.
ARTEMIS is available at GitHub: https://github.com/david-bogdan-r/ARTEMIS
Thanks to Davyd Bohdan, Janusz Bujnicki, & International Institute of Molecular and Cell Biology in Warsaw!

#rna#dna#3d#science#bioinformatics#structuralbiology#research#innovation#structure#topology#sequence#superposition#alignment
8 notes
·
View notes
Text
my ocd has latched onto. the work i do at my job
5 notes
·
View notes
Text
5 notes
·
View notes
Text
Real-World Applications of Gene Expression Profiling and Pathway Enrichment in Cancer Research
Cancer remains one of the leading causes of death worldwide, prompting ongoing research to uncover its underlying mechanisms and develop effective treatments. Among the numerous tools available to cancer researchers, gene expression profiling and pathway enrichment analysis have emerged as powerful methods for understanding tumor biology, identifying therapeutic targets, and improving patient outcomes. This article explores the real-world applications of gene expression profiling in cancer research, highlighting how these techniques are reshaping the landscape of cancer diagnosis and treatment.

To explore more about applications of Gene Expression Profiling and Pathway Enrichment Analysis you can visit Gene-omics.
If you want to learn about the genomics from the beginning, you can enroll for the Genome Analysis Bootcamp Demo Session here- https://edgeneomics.com/genome-analysis-bootcamp/
To learn Gene Expression Analysis, you may visit https://edgeneomics.com/gene-expression-with-r-masterclass/
0 notes
Text
queen belfast v imperial was so good oh my gosh
but wow i really love both teams (wish they both somehow could’ve gone through 😢 could basically hear my heartbeat during the tiebreaker)
0 notes
Text
AMPure XP vs. HighPrep PCR: Which DNA Cleanup Solution Delivers Better Performance?
Next-generation sequencing (NGS), PCR, and other molecular biology workflows rely on effective DNA cleanup solutions to ensure high-quality results. Among the most popular choices are AMPure XP and HighPrep PCR, both of which utilize SPRI (Solid Phase Reversible Immobilization) bead-based purification. But which one performs better? And which is the best choice for your lab? In this article, we provide a detailed performance comparison of AMPure XP and HighPrep PCR to help you make an informed decision.
Why DNA Cleanup Matters
DNA cleanup is a critical step in molecular biology workflows, as contaminants such as salts, primers, nucleotides, and enzymes can interfere with downstream applications like PCR, qPCR, and sequencing. SPRI bead-based purification has become the gold standard for DNA cleanup due to its ability to efficiently remove impurities while preserving high DNA recovery rates.
AMPure XP vs. HighPrep PCR: Head-to-Head Comparison
To compare the two purification systems, we evaluated them based on key performance factors, including DNA recovery efficiency, purity, ease of use, compatibility, and cost-effectiveness.
1. DNA Recovery Efficiency
The primary goal of any DNA cleanup method is to retain as much high-quality DNA as possible while removing contaminants.
AMPure XP: Delivers consistently high DNA recovery rates (>90%) and is widely used in NGS and PCR workflows.
HighPrep PCR: Also provides excellent recovery (>90%) and has been validated for use in various molecular biology applications.
Winner: Tie – Both kits deliver high DNA yield and recovery.
2. DNA Purity and Contaminant Removal
Ensuring pure DNA is essential for high-performance downstream applications.
AMPure XP: Effectively removes contaminants, but some reports indicate occasional ethanol carryover, which can affect sensitive reactions.
HighPrep PCR: Equally efficient in removing contaminants but features an optimized protocol to minimize ethanol carryover.
Winner: HighPrep PCR – Slightly better ethanol carryover prevention.
3. Ease of Use & Protocol Simplicity
Both purification kits follow a standard bind-wash-elute protocol, but minor differences can impact usability.
AMPure XP: Standard 3-step workflow with a 30-minute processing time.
HighPrep PCR: Offers an optimized, shorter workflow that reduces processing time by 10-15%.
Winner: HighPrep PCR – Faster and slightly more efficient workflow.
4. Compatibility with Manual and Automated Workflows
Many labs require flexibility between manual processing and high-throughput automation.
AMPure XP: Fully compatible with liquid-handling automation systems.
HighPrep PCR: Designed for both manual and automated workflows, with high adaptability for robotics.
Winner: Tie – Both support automation effectively.
5. Cost-Effectiveness
Budget constraints are a major factor in choosing DNA cleanup solutions.
AMPure XP: Premium-priced, making it a costly option for labs running high-throughput workflows.
HighPrep PCR: Offers a more affordable alternative with comparable performance, making it a cost-effective solution for research labs.
Winner: HighPrep PCR – More budget-friendly without compromising performance.
Final Verdict: Which One Should You Choose?
If your lab prioritizes high recovery, purity, and automation compatibility, both AMPure XP and HighPrep PCR are excellent choices. However, HighPrep PCR stands out due to its better cost-effectiveness, faster workflow, and improved ethanol carryover prevention, making it the ideal choice for budget-conscious labs looking for reliable performance.
Where to Get HighPrep PCR?
MagBio Genomics provides a cost-effective, high-performance alternative to AMPure XP with HighPrep PCR.
Ready to optimize your DNA cleanup workflow? Learn more and order your kit today at MagBio Genomics.
0 notes