#two-cell homologous recombination
Text
If organisms without respiratory systems are our ancestors
If organisms without respiratory systems are our ancestors
Why then do organisms without respiratory systems not exist today
Can alien organisms like Luca still exist today
The Last Universal Common Ancestor (LUCA) is a hypothetical cell that is believed to have lived between 3.6 and 4.3 billion years ago, and from which all life evolved. There is no fossil evidence of LUCA, but the biochemical similarities of all current life strongly support its existence.
Some believe that LUCA may have lived in hydrothermal vents deep underground, and that it may have had a somewhat "alien" lifestyle. However, others believe that there may not be a single LUCA, but rather an indefinable evolutionary starting point for life.
While no fossil evidence of the LUCA exists, the detailed biochemical similarity of all current life (divided into the three domains) makes its existence widely accepted by biochemists. Its characteristics can be inferred from shared features of modern genomes.
Looking for LUCA, the Last Universal Common Ancestor
NASA Astrobiology (.gov)
https://astrobiology.nasa.gov › news › looking-for-luca...
30 Mar 2017
Ancient genes
Previous studies of LUCA looked for common, universal genes that are found in all genomes, based on the assumption that if all life has these genes, then these genes must have come from LUCA. This approach has identified about 30 genes that belonged to LUCA, but they’re not enough to tell us how or where it lived. Another tactic involves searching for genes that are present in at least one member of each of the two prokaryote domains, archaea and bacteria. This method has identified 11,000 common genes that could potentially have belonged to LUCA, but it seems far-fetched that they all did: with so many genes LUCA would have been able to do more than any modern cell can.
Bill Martin and his team realized that a phenomenon known as lateral gene transfer (LGT) was muddying the waters by being responsible for the presence of most of these 11,000 genes. LGT involves the transfer of genes between species and even across domains via a variety of processes such as the spreading of viruses or homologous recombination that can take place when a cell is placed under some kind of stress.
A growing bacteria or archaea can take in genes from the environment around them by ‘recombining’ new genes into their DNA strand. Often this newly-adopted DNA is closely related to the DNA already there, but sometimes the new DNA can originate from a more distant relation. Over the course of 4 billion years, genes can move around quite a bit, overwriting much of LUCA’s original genetic signal. Genes found in both archaea and bacteria could have been shared through LGT and hence would not necessarily have originated in LUCA.
Knowing this, Martin’s team searched for ‘ancient’ genes that have exceptionally long lineages but do not seem to have been shared around by LGT, on the assumption that these ancient genes should therefore come from LUCA. They laid out conditions for a gene to be considered as originating in LUCA. To make the cut, the ancient gene could not have been moved around by LGT and it had to be present in at least two groups of archaea and two groups of bacteria.
“While we were going through the data, we had goosebumps because it was all pointing in one very specific direction,” says Martin.
Once they had finished their analysis, Bill Martin’s team was left with just 355 genes from the original 11,000, and they argue that these 355 definitely belonged to LUCA and can tell us something about how LUCA lived.
Such a small number of genes, of course, would not support life as we know it, and critics immediately latched onto this apparent gene shortage, pointing out that essential components capable of nucleotide and amino acid biosynthesis, for example, were missing. “We didn’t even have a complete ribosome,” admits Martin.
However, their methodology required that they omit all genes that have undergone LTG, so had a ribosomal protein undergone LGT, it wouldn’t be included in the list of LUCA’s genes. They also speculated that LUCA could have gotten by using molecules in the environment to fill the functions of lacking genes, for example molecules that can synthesize amino acids. After all, says Martin, biochemistry at this early stage in life’s evolution was still primitive and all the theories about the origin of life and the first cells incorporate chemical synthesis from their environment.
What those 355 genes do tell us is that LUCA lived in hydrothermal vents. The Düsseldorf team’s analysis indicates that LUCA used molecular hydrogen as an energy source. Serpentinization within hydrothermal vents can produce copious amounts of molecular hydrogen. Plus, LUCA contained a gene for making an enzyme called ‘reverse gyrase’, which is found today in extremophiles existing in high-temperature environments including hydrothermal vents.
Friends, Mars is the twin sister of our Earth.
My favourite planet.
I have written articles about Mars many times.
This time I have written about Mars in the Solar System Project.
You decide whether this article of mine will give the right direction to the coming generation or not.
Till then allow me to start about Jupiter.
Translate Hindi
बिना श्वसन प्रणाली के जीव अगर हमारे पूर्वज है
क्यों फिर श्वसन प्रणाली बिन जीव आज अस्तित्व में है ही नहीं
क्या लूका जैसी एलियन जीव आज भी हो सकता है
अंतिम सार्वभौमिक सामान्य पूर्वज (LUCA) एक काल्पनिक कोशिका है जिसके बारे में माना जाता है कि यह 3.6 से 4.3 बिलियन वर्ष पहले रहती थी और जिससे सभी जीवन विकसित हुए। LUCA का कोई जीवाश्म साक्ष्य नहीं है, लेकिन सभी मौजूदा जीवन की जैव रासायनिक समानताएँ इसके अस्तित्व का दृढ़ता से समर्थन करती हैं।
कुछ लोगों का मानना है कि LUCA शायद भूमिगत हाइड्रोथर्मल वेंट में रहता था और इसकी जीवनशैली कुछ हद तक "एलियन" रही होगी। हालाँकि, अन्य लोगों का मानना है कि एक भी LUCA नहीं हो सकता है, बल्कि जीवन के लिए एक अनिश्चित विकासवादी प्रारंभिक बिंदु हो सकता है।
जबकि LUCA का कोई जीवाश्म साक्ष्य मौजूद नहीं है, सभी मौजूदा जीवन (तीन डोमेन में विभाजित) की विस्तृत जैव रासायनिक समानता इसके अस्तित्व को जैव रसायनज्ञों द्वारा व्यापक रूप से स्वीकार करती है। इसकी विशेषताओं का अनुमान आधुनिक जीनोम की साझा विशेषताओं से लगाया जा सकता है।
LUCA, अंतिम सार्वभौमिक सामान्य पूर्वज की खोज
NASA एस्ट्रोबायोलॉजी (.gov)
https://astrobiology.nasa.gov › news › looking-for-luca...
30 मार्च 2017
प्राचीन जीन
LUCA के पिछले अध्ययनों में सभी जीनोम में पाए जाने वाले सामान्य, सार्वभौमिक जीन की तलाश की गई थी, इस धारणा के आधार पर कि यदि सभी जीवन में ये जीन हैं, तो ये जीन LUCA से आए होंगे। इस दृष्टिकोण ने लगभग 30 जीन की पहचान की है जो LUCA से संबंधित हैं, लेकिन वे हमें यह बताने के लिए पर्याप्त नहीं हैं कि यह कैसे या कहाँ रहता था। एक अन्य रणनीति में उन जीनों की खोज करना शामिल है जो दो प्रोकैरियोट डोमेन, आर्किया और बैक्टीरिया में से प्रत्येक के कम से कम एक सदस्य में मौजूद हैं। इस पद्धति ने 11,000 सामान्य जीन की पहचान की है जो संभावित रूप से LUCA से संबंधित हो सकते हैं, लेकिन ऐसा लगता है कि वे सभी ऐसा करते हैं: इतने सारे जीन के साथ LUCA किसी भी आधुनिक कोशिका से अधिक करने में सक्षम होगा।
बिल मार्टिन और उनकी टीम ने महसूस किया कि पार्श्व जीन स्थानांतरण (LGT) के रूप में जानी जाने वाली घटना इन 11,000 जीनों में से अधिकांश की उपस्थिति के लिए जिम्मेदार होने के कारण पानी को गंदा कर रही थी। LGT में विभिन्न प्रकार की प्रक्रियाओं जैसे वायरस का प्रसार या समजातीय पुनर्संयोजन के माध्यम से प्रजातियों के बीच और यहाँ तक कि डोमेन में जीन का स्थानांतरण शामिल है जो तब हो सकता है जब किसी कोशिका को किसी प्रकार के तनाव में रखा जाता है।
बढ़ते बैक्टीरिया या आर्किया अपने डीएनए स्ट्रैंड में नए जीन को 'पुनर्संयोजित' करके अपने आस-पास के वातावरण से जीन ले सकते हैं। अक्सर यह नया अपनाया गया डीएनए पहले से मौजूद डीएनए से बहुत करीब से संबंधित होता है, लेकिन कभी-कभी नया डीएनए अधिक दूर के रिश्तेदार से उत्पन्न हो सकता है। 4 बिलियन वर्षों के दौरान, जीन काफी हद तक इधर-उधर हो सकते हैं, जो LUCA के मूल आनुवंशिक संकेत को अधिलेखित कर सकते हैं। आर्किया और बैक्टीरिया दोनों में पाए जाने वाले जीन LGT के माध्यम से साझा किए जा सकते थे और इसलिए जरूरी नहीं कि वे LUCA में उत्पन्न हुए हों। यह जानते हुए, मार्टिन की टीम ने ऐसे 'प्राचीन' जीन की खोज की, जिनकी वंशावली असाधारण रूप से लंबी है, लेकिन ऐसा नहीं लगता कि LGT द्वारा साझा की गई है, इस धारणा पर कि ये प्राचीन जीन इसलिए LUCA से आने चाहिए। उन्होंने LUCA में उत्पन्न होने वाले जीन के लिए शर्तें रखीं। कट बनाने के लिए, प्राचीन जीन LGT द्वारा इधर-उधर नहीं किया जा सकता था और इसे कम से कम दो समूहों के आर्किया और दो समूहों के बैक्टीरिया में मौजूद होना चाहिए था। मार्टिन कहते हैं, "जब हम डेटा देख रहे थे, तो हमारे रोंगटे खड़े हो गए क्योंकि यह सब एक बहुत ही विशिष्ट दिशा की ओर इशारा कर रहा था।" जब उन्होंने अपना विश्लेषण पूरा कर लिया, तो बिल मार्टिन की टीम के पास मूल 11,000 में से केवल 355 जीन बचे थे, और उनका तर्क है कि ये 355 निश्चित रूप से LUCA के थे और हमें इस बारे में कुछ बता सकते हैं कि LUCA कैसे रहता था। जीन की इतनी कम संख्या, निश्चित रूप से, जी��न का समर्थन नहीं करेगी जैसा कि हम जानते हैं, और आलोचकों ने तुरंत इस स्पष्ट जीन की कमी पर ध्यान दिया, यह इंगित करते हुए कि न्यूक्लियोटाइड और अमीनो एसिड जैवसंश्लेषण में सक्षम आवश्यक घटक, उदाहरण के लिए, गायब थे। मार्टिन ने स्वीकार किया, "हमारे पास एक पूरा राइबोसोम भी नहीं था।" हालांकि, उनकी कार्यप्रणाली के लिए आवश्यक था कि वे उन सभी जीनों को छोड़ दें जो LTG से गुजर चुके हैं, इसलिए यदि कोई राइबोसोमल प्रोटीन LGT से गुजरा होता, तो उसे LUCA के जीन की सूची में शामिल नहीं किया जाता। उन्होंने यह भी अनुमान लगाया कि LUCA पर्यावरण में अणुओं का उपयोग करके जीन की कमी को पूरा कर सकता है, उदाहरण के लिए अणु जो अमीनो एसिड को संश्लेषित कर सकते हैं। आखिरकार, मार्टिन कहते हैं, जीवन के विकास में इस प्रारंभिक चरण में जैव रसायन अभी भी आदिम था और जीवन की उत्पत्ति और पहली कोशिकाओं के बारे में सभी सिद्धांत अपने पर्यावरण से रासायनिक संश्लेषण को शामिल करते हैं। वे 355 जीन हमें यह बताते हैं कि LUCA हाइड्रोथर्मल वेंट में रहता था। डसेलडोर्फ टीम के विश्लेषण से पता चलता है कि LUCA ने ऊर्जा स्रोत के रूप में आणविक हाइड्रोजन का उपयोग किया। हाइड्रोथर्मल वेंट के भीतर सर्पेंटिनाइजेशन से आणविक हाइड्रोजन की प्रचुर मात्रा उत्पन्न हो सकती है। साथ ही, LUCA में ‘रिवर्स गाइरेस’ नामक एंजाइम बनाने के लिए एक जीन होता है, जो आज हाइड्रोथर्मल वेंट सहित उच्च तापमान वाले वातावरण में मौजूद एक्सट्रीमोफाइल्स में पाया जाता है।
दोस्तों वैसे मंगल हमारा पृथ्वी का जुड़वा बहने है
मेरा खास फेवोरिट प्लैनेट
काफी बार मैं मंगल के बारे में आर्टिकल लिखा हूँ
इस बार सौरमंडल प्रोजेक्ट के अंदर की मंगल को मैं लिखा हूँ
मेरा यह आर्टिकल आनेवाला पिढ़ी को सही दिशा देगी या नहीं यह आप सोचिए
तब तक मुझें वृहस्पति के बारे में शुरू करने की इजाजत दीजिए
0 notes
Text
Ovarian Cancer Drugs: Fighting a Silent Killer

Ovarian cancer is often referred to as the "silent killer" due to its difficulty in detection at an early stage. While ovarian cancer accounts for only about 3% of all cancers in women, it causes more deaths than any other cancer of the female reproductive system. Fortunately, major advances have been made in ovarian cancer drugs that are helping more women survive this disease. This article provides an overview of some key drugs used to treat ovarian cancer.
Platinum-Based Chemotherapy Drugs
Platinum-based chemotherapy drugs, such as carboplatin and cisplatin, have been the standard treatments for ovarian cancer for decades. These drugs work by cross-linking DNA strands in cancer cells, preventing them from replicating. Though extremely toxic, platinum drugs are effective at destroying fast-growing cancer cells. In the first-line treatment of ovarian cancer, carboplatin is usually administered along with paclitaxel or another chemotherapy drug to increase effectiveness. While platinum drugs are not curative on their own, they significantly improve survival rates when used as part of initial treatment.
angiogenesis inhibitors disrupt the formation of new blood vessels that tumors need to grow. One such drug used to treat ovarian cancer is bevacizumab (Avastin). As an angiogenesis inhibitor, bevacizumab starves tumors of nutrients and oxygen by blocking vascular endothelial growth factor (VEGF). Studies have found bevacizumab improves progression-free survival when combined with chemotherapy as a first-line or maintenance treatment for ovarian cancer. The drug is also being examined as a potential treatment for recurrent disease. Side effects can include high blood pressure, bleeding, problems wound healing and gastrointestinal perforations. Careful monitoring is required with bevacizumab treatment.
PARP Inhibitors: A New Target
Poly (ADP-ribose) polymerase, or PARP, inhibitors are an exciting new class of drugs for ovarian cancer. PARP is an enzyme involved in DNA repair. PARP inhibitors work by blocking cancer cells already deficient in homologous recombination repair, like those with BRCA gene mutations, from repairing DNA damage. This leaves the cancer vulnerable to cell death. In 2017, the FDA approved the PARP inhibitor olaparib (Lynparza) for the maintenance treatment of recurrent ovarian cancer patients with a BRCA mutation who have responded to platinum-based chemotherapy. More recently, it was approved for germline BRCA-mutated advanced ovarian cancer after three or more lines of chemotherapy. Rubraca (rucaparib) and Zejula (niraparib) are two other PARP inhibitors approved to treat ovarian cancer. Ongoing research continues to explore the potential of PARP inhibitors in different treatment settings and biomarker populations. Side effects can include fatigue, nausea, vomiting, diarrhea and low blood cell counts.
0 notes
Text
Genetics: Meiosis
Meiosis is a form of cell division to form gametes (sex cells). The two main functions is to form haploid cells with half the normal chromosome number and to re-arrange the chromosomes with a novel recombination of genes. An aspect of meiosis that makes it so unique, is that it goes through two divisions. This forms four unique daughter cells and aids in genetic variety.
Summary:
In Prophase I, the DNA condenses and matches up with homologous pair (chromosome pair
from mother matches with same chromosome pair from father). Crossing-over takes place.
Mother and father chromosomes overlap and swap leading to recombinant chromosomes
In Metaphase I, recombinant chromosomes line up at middle.
Anaphase I, chromosomes move to opposite sides of the cell.
Telophase I, new nuclei form around chromosomes.
Cytokinesis, cell splits into two cells. Each has 23 pairs of chromosomes (46 chromatids), 23
centromeres.
Prophase II, chromosomes condense, no crossing-over.
Metaphase II, single line of 23 chromosome pairs at middle of each daughter cell. (Occurs BOTH
the cells created in cytokinesis)
Anaphase II, chromatids (single) get pulled to opposite ends of each of the daughter cells, 23
chromatids to each end.
Telophase II, new nuclear membranes form (2 cells with two nuclei forming in each)
0 notes
Text
Are There Any New Cholangiocarcinoma Treatment options?
If you have been diagnosed with cholangiocarcinoma, you may be asking if there are any new treatments for this disease. The good news is that you have a number of alternatives, including surgical resection, FGFR inhibitors, and PARP inhibitors. However, you should be aware that there are a few potential hazards associated with these treatment approaches.
Cholangiocarcinoma is rare cancer that develops in the bile duct epithelial cells. It might be localized or diffuse, affecting the biliary tree at any point. The only potentially curable treatment is surgical resection.
The history, physical examination, and test results are used to make a cholangiocarcinoma diagnosis. Patients are assessed after resection to determine their prognosis. Surgical resection is usually successful. However, recurrence is possible following resection.
Depending on the location of the tumor, there are numerous resection options. The most common treatment for intrahepatic cholangiocarcinomas is liver resection. Distal/perihilar cholangiocarcinomas necessitate bile duct resection. Preoperative liver drainage is recommended for patients undergoing hepatic resections. Partial liver resections are frequently paired with total caudate lobe excision.
Partial hepatectomy, bile duct resection, and extensive hepatic resection are the most common treatments for perihilar cholangiocarcinomas. Overall survival is reduced in distal and perihilar cholangiocarcinomas. They are also more prone to perineural invasion.
FGFR inhibitors as a potential therapeutic option for cholangiocarcinoma have developed. While standard treatments have a bad prognosis for this condition, tailored therapy may be a viable choice.
Several FGFR inhibitors are now in clinical trials. Debit-1347 and pemigatinib are two examples. Debio-1347 is a strong multikinase inhibitor that can be used orally. It has been demonstrated that it specifically targets FGFR-deregulated cancers. Pemigatinib was found to be beneficial in individuals with advanced cholangiocarcinoma who had FGFR2 rearrangements in phase II trials.
Pemigatinib is an FGFR1-3 inhibitor authorized for oral use by the US Food and Drug Administration (FDA) in April 2020. The European Commission is expected to approve the project in March 2021. A phase II trial of this medication for metastatic cholangiocarcinoma was also conducted.
Infigratinib, another FGFR inhibitor, is being studied as the first-line therapy for FGFR2-fusions in unresectable cholangiocarcinoma. A phase III investigation is now underway.
Pemigatinib has been found to be a highly effective and selective FGFR1 and FGFR2 inhibitor. It is a viable therapy for FGFR-driven malignancies due to its strong affinities for both FGFR1 and FGFR2, as well as its vast hydrogen bond network.
PARP inhibitors are a unique family of chemotherapeutic medicines that have shown potential clinical activity in a variety of solid cancers. The Food and Drug Administration has currently approved three PARP inhibitors, and several other medications are in development. They work with people with BRCA-mutant malignancies. Despite the good clinical results, more research is needed to discover which patients will benefit the most from this therapy. Meanwhile, a number of clinical trials involving these compounds are exploring new medication combinations.
PARP (poly ADP-ribose polymerase) is an enzyme that is essential in the repair of DNA single-strand breaks. BRCA gene mutations cause genomic instability, rendering the homologous recombination (HR) mechanism inefficient. PARP inhibition may be useful in decreasing the mutational burden in HRD-associated malignancies by inhibiting the cells' ability to repair DSBs.
Preclinical and early clinical results suggest that PARP inhibition may be beneficial in restoring the deficit caused by an IDH1 mutation. These mutations cause isocitrate to be converted into 2-hydroxyglutarate, which enhances tumor cell proliferation. As a result, the PI3k pathway, a prevalent driver of tumor development, is frequently constitutively activated.
The neurotrophic tyrosine receptor kinases (NTRK) genes are necessary for nerve cell formation and maintenance. They also aid in the development of certain types of cancers. When one of these genes is rearranged, a fusion is formed and produced as a chimeric protein.
Sarcomas, colon cancer, secretary breast carcinoma, non-small cell lung cancer, cholangiocarcinoma, and melanoma all include NTRK fusions. They are, nevertheless, uncommon. This makes determining whether a patient's tumor has an altered NTRK gene difficult.
There are several approaches for screening for a rearranged NTRK. The majority of oncologists choose next-generation sequencing. However, in some circumstances, IHC is preferable.
NTRK fusions have been found in patients with a range of malignancies, with non-small cell lung cancer or sarcoma being the most common. NTRK fusion cancers are classified into two types: rare and common.
1 note
·
View note
Text
Cancer Epigenetic Regulation
Knowing the mechanisms underlying epigenetic regulation will help you better comprehend how cancer arises and how to treat it. The change of chromatin, the genetic material, and the addition of nucleic acids are two aspects of the epigenetic regulatory process. These two procedures are crucial in explaining why some individuals develop cancer and others do not.
MYC controls a wide range of gene programs in healthy cells, according to molecular research on the protein's function in cancer. It is crucial for controlling the growth of malignancies. Despite the lack of a clear causal relationship between MYC and oncogenic reprogramming, the findings of these investigations suggest that MYC is essential for both tumor initiation and growth regulation. MYC is very important for sustaining pluripotency and apoptosis. It plays a significant role in oncogenic reprogramming as well.
MYC is a key player in the growth of basal-like breast cancers, according to earlier studies. This work demonstrated that overexpression of MYC promotes the reactivation of a transcriptional pathway linked to pluripotency in the luminal epithelial cells of mammary tissue. Given that basal-like breast cancers are distinguished by the overexpression of the oncogene MYC, this pattern of expression could encourage carcinogenesis.
Numerous cancer treatments concentrate on epigenetic control. Epi-drugs are the name given to these medications. These medications have demonstrated encouraging anti-tumor effects in clinical studies. They have not yet received clinical use approval, nevertheless.
A cell's epigenetic state affects both its ability to survive and grow. Epigenetic markers may alter in status in response to environmental or physiological changes. Cell death may result if the alterations are not adaptive.
DNA methylation, histone modification, and non-coding RNA regulation are the three main categories of epigenetic alterations. Through interactions with other molecular elements, these alterations are reversible and re-regulated. For the onset and spread of cancer, these alterations are crucial.
The most frequent epigenetic change, DNA methylation, affects over 70% of the CpG dinucleotides in the mammalian genome. Through the covalent addition of methyl groups to the cytosine pyrimidine ring's 5 position, DNA methylation causes gene silence.
Epigenetic regulation is an intriguing target among the many that might be used in cancer treatments. It is dynamic, reversible, and subject to external influences. It comprises non-coding RNA (ncRNA) control, DNA methylation, and chromatin changes. The significance of these changes in cancer has been the subject of several research, and it seems likely that they contribute to carcinogenesis.
BET proteins are important for chromatin remodeling and have a role in gene expression. For the purpose of regulating RNA polymerase II dependent transcription, they assemble complexes with other proteins, such as HDACs. They are also recognized as readers of histone acetyl-lysine. BETs have also been demonstrated to control homologous recombination-mediated DNA damage repair.
The functions of BETs in cancer have been studied by academics in recent years. The extra-terminal peptide SLUG and the bromodomain-containing protein BRD4 are two of the BET family's best-studied members. It was discovered that BRD4 and SLUG cooperate to control BRCAness induction treatment in breast cancer cells. Additionally, they have been recognized as side effects of other oncogenes.
Chemokines have a key role in a number of crucial tumor formation processes, including the attraction of immune cells to the tumor microenvironment (TME). Chemokines are also known to influence the proliferation and differentiation of cancer cells. They may also encourage angiogenesis, inflammation, and metastasis. The cytokines are inhibited in many malignancies by epigenetic changes. These alterations include interactions with other molecular elements, DNA methylation, histone changes, chemokine receptor suppression, and histone modifications.
Inflammatory cells support angiogenesis and the growth of cancer cells in the tumor microenvironment. Additionally, in order to promote tumor development, cancer cells manipulate the immunological microenvironment. By converting the structure of the healthy tissue into an inflammatory milieu, they do this.
One of the most significant chemokines that affect the tumor microenvironment is CXCL12. In the tumor microenvironment, a variety of cell types, including fibroblasts, innate lymphoid cells, and tumor-associated macrophages, produce CXCL12. High levels of CXCL12 expression are linked to a poor outcome in breast cancer.
Combining epi-drugs with traditional cancer treatments can be a potent tool in the arsenal. Epi-drugs have been shown to have synergistic benefits in the treatment of different cancers in a number of trials. Epi-drugs, in contrast to conventional anti-cancer medications, can target epigenetic alterations and hence improve tumor inhibition. They are a perfect fit for multi-drug regimens since they also have the advantage of being targeted at a particular area of the body.
The effectiveness of epi-drugs in the management of malignancies is still up for debate. They have been demonstrated to cause epigenetic alterations that may eventually result in acquired resistance. Clinical studies for these substances are still being conducted. Thus, the demand for epi-drugs with a clearly established safety profile exists.
0 notes
Text
Macvector protein sequence

Overall sequence similarity with the mouse protein is shown on the right. The percent similarity of the SAM domains and other regions to the corresponding regions of the mouse protein is shown. (D) Schematic comparison of the amino acid sequences for mouse, rat, human, chick and zebrafish mr-s proteins. Branch lengths reflect the mean number of substitutions per site. Amino acid sequences were analyzed by the neighbor-joining method in MacVector 7.2. (C) Phylogenetic tree of SAM domain-containing proteins. The sites that were targeted for mutagenesis are indicated by arrows. Conserved amino acid residues are shown with a dark shadow and functionally similar residues are shown with a light shadow. (B) Alignment of SAM domain sequences for SAM domain-containing proteins. The underline indicates a putative polyadenylation termination signal. Boxed amino acids are the SAM domain sequence and the dashed box indicates a putative nuclear localization signal. (A) mr-s nucleotide and amino acids sequences. In addition, the finding of rare heterozygous QRX sequence changes in three individuals with retinal degeneration raises the possibility that QRX may be involved in disease pathogenesis.Mr-s nucleotide and amino acid sequences. These results indicate that Qrx may be involved in modulating photoreceptor gene expression. Nonetheless, a 5.8 kb upstream region of human QRX is capable of directing expression in presumptive photoreceptor precursor cells in transgenic mice. Qrx is present in the bovine and human genomes, but appears to be absent from the mouse genome. As predicted from the amino acid sequence homology, recombinant DiTG catalyzed. QRX synergistically increases the transactivating function of the photoreceptor transcription factors Crx and NRL and it physically interacts with CRX. protein ERp60, a PDI isoform found in the lumen of endo- plasmic reticulum. Although Qrx and Rx/Rax show similar DNA binding properties in vitro, the two proteins demonstrate distinct target selectivity and functional behavior in promoter activity assays. Its homeodomain is nearly identical to that of Rx/Rax, a transcription factor that is essential for eye development, but it shares only limited homology elsewhere. Qrx is preferentially expressed in both the outer and inner nuclear layers of the retina. Ī novel paired-like homeobox gene, designated as Qrx, was identified by a yeast one-hybrid screen using the bovine Rhodopsin promoter Ret-1 DNA regulatory element as bait. Six microliters (20 ng/ml) purified Scc1 or Scc1-P in H100 GIBBS sampling option of the MACAW program (Schuler et al., 1991 was added to. Multiple sequenceScc1 cleavage-competent yeast extracts after overexpression of alignment construction and analysis, with statistical evaluation ofEsp1 and control extracts were obtained as described (Uhlmann et the significance of motif conservation, were performed using theal., 1999). to the MacVector 6.5. Database screening for amino acid patterns was performed using the PATTINPROT pro-Cleavage of Purified Scc1 in Yeast Extracts gram at the NPS2 server (Combet et al., 2000). The L-Zip motif sequence and the confirmation of the two helices were found by submitting the nad4L amino acid sequences we obtained for several schistosome spp. the PHI-BLAST program (Zhang et al., 1998). Additional database searches combining BLASTP with a pattern analysis were performed usingmetaphase-like phosphorylated Scc1. This protocol yielded 25 mg of purified Scc1 or purified BLAST program (Altschul et al., 1997). The final eluate was dialyzed against buffer H100 and concentrated bytion by cleaving other proteins or by a different mechaCell 384 ultrafiltration. Whether Esp1 performs this func- as Hsp70 from the insect host cells by mass spectrometry. 96a Ciosk et al., 1998 contaminating protein that copurified with Scc1 and was identified Kumada et al., 1998). To access GenBank and its related retrieval and analysis services, go to the NCBI home page at: Complete bimonthly releases and daily updates of the GenBank database are available by FTP. BLAST provides sequence similarity searches of GenBank and other sequence databases. GenBank is accessible through NCBI's retrieval system, Entrez, which integrates data from the major DNA and protein sequence databases along with taxonomy, genome mapping, protein structure and domain information, and the biomedical journal literature via PubMed. Daily data exchange with the EMBL Data Library in the UK and the DNA Data Bank of Japan helps ensure worldwide coverage. Most submissions are made using the BankIt (web) or Sequin program and accession numbers are assigned by GenBank staff upon receipt. GenBank (R) is a comprehensive database that contains publicly available DNA sequences for more than 140 000 named organisms, obtained primarily through submissions from individual laboratories and batch submissions from large-scale sequencing projects.

0 notes
Text
Binary fission diagram

Binary fission diagram full size#
The nucleus of the parent cell splits into a daughter nucleus and migrates into the daughter cell.
Binary fission diagram full size#
First it produces a small protuberance on the parent cell that grows to a full size and forms a bud. Most yeasts reproduce asexually by an asymmetric division process called budding. Yeast size can vary greatly depending on the species, typically measuring 3-4 µm in diameter. Yeast are unicellular (some are multicellular) eukaryotic micro-organisms belonging to the kingdom fungi. Cytokinesis occurs immediately after mitosis. It corresponds to the separation of the daughter nuclei into two daughter cells. Karyokinesis is usually followed by Cytokinesis.Ĭytokinesis is the process of the division of the cytoplasm. It corresponds to the separation of the daughter chromosomes into two daughter nuclei. Karyokinesis is the process of the division of the nucleus. This leads to the formation of the two daughter Amoebae cell having a nucleus and its own cell organelles. After the nucleus has divided into two, the process of Cytokinesis takes place in which the cytoplasm in the mother cell divides into two daughter cells. In this process, the nucleus of the Amoeba first divides to form two daughter nuclei by the process of Karyokinesis. The genetic material is also equally partitioned therefore the daughter cells are genetically identical to each other and the parent cell. After replicating its genetic material through mitotic division, the cell divides into two equal sized daughter cells. Amoeba reproduces by the common asexual reproduction method called binary fission. Binary fission in AmoebaĪmoeba is a shapeless tiny unicellular organism that has a porous cell membrane which encloses the cell organelles and cytoplasm. Binary fission is found in unicellular organisms like Amoeba, Paramaecium and Euglena, to name and few. It is a form of nuclear division.īinary fission and budding are two common method of asexual reproduction. Mitosis is the process by which a cell, which has previously replicated each of its chromosomes, separates the chromosomes in its cell nucleus into two identical sets of chromosomes, each set will have its own new nucleus. Amitosis is the process by which a cell directly separates, as the nucleus and cytoplasm are directly cut in two. New organisms are produced in rapid multiplication by the process of amitotic or mitotic divisions. The offspring will be the exact genetic copies of the parent. Asexual reproduction is a mode of reproduction by which offspring arise from a single parent, and inherit the genes of that parent only. Many plants and fungi reproduce asexually as well. Asexual reproductionĪsexual reproduction is the primary form of reproduction for single-celled organisms such as the archaea, bacteria, and protists. During meiosis, the chromosomes of each pair usually cross over to achieve homologous recombination that helps produce genetic diversity when cells divide in meiosis. Here are two main processes during sexual reproduction in eukaryotes: meiosis, involving the halving of the number of chromosomes and fertilisation, involving the fusion of two gametes and the restoration of the original number of chromosomes. Sexual reproduction is the primary method of reproduction for the vast majority of macroscopic organisms, including almost all animals and plants. Reproduction takes place sexually and asexually. Our objective is to study using the prepared slides Īll living things produce their own kind through the process called reproduction.

0 notes
Text
Unit 4b: How Meiosis Works
During meiosis, one cell divides twice, thus producing four cells, each with ½ the number of chromosomes. Therefore, each gamete has only ½ the genetic information. This makes the gametes haploid (as opposed to diploid).
Meoisis can be divided into 9 stages. The first time that the cell divides is meiosis I, and the second time it divides is meiosis II.
Background
A pair of homologous chromosomes is a set of one maternal and one paternal chromosome. These two chromosomes pair up with each other inside the cell during fertilization.
The two homologous chromosomes have the same:
gene sequence
gene loci
chromosomal length
centromere location
(But they differ in alleles.)
Each maternal chromosome has a corresponding paternal chromosome, and they pair up during meiosis.
The sex chromosomes (the 23rd pair) are homologous in females (XX) but not in males (XY).
Non-homologous chromosomes are called heterologous chromosomes.
Here is the first homologous pair of chromosomes (called Chromosome 1):
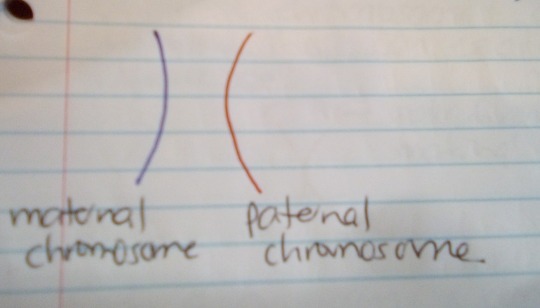
-
MEIOSIS I
Interphase
The DNA in the cell is copied (replicated). This results in 2 identical full sets of chromosomes.
Here are the original and duplicate of Chromosome 1 (1a is the original, and 1b is the duplicate):

There are two centrosomes outside of the nucleus, each containing a pair of centrioles. The centrosomes are essential for the process of cell division.
During interphase, microtubules extend from these centrosomes.
Prophase I
The copied chromosomes condense into X-shaped structures that can easily be seen under a microscope. The maternal chromosome pairs up with its duplicate, and the paternal chromosome pairs up with its duplicate.

Each chromosome is composed of 2 sister chromatids with identical genetic information.
The chromosomes now pair up so that the maternal pair & paternal pair from Chromosome 1 are together, etc.
The pairs of chromosomes may then exchange bits of DNA in a process called recombination or “crossing over”.
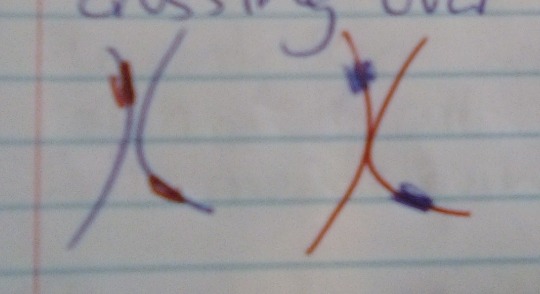
So now the 2 maternal chromosomes (in blue) are no longer completely identical, and neither are the 2 paternal chromosomes (in red).
At the end of this phase, the membrane around the cell nucleus dissolves away. This releases the chromosomes.
The meiotic spindle consists of microtubules and other proteins. It extends across the cell between the centrioles.
Metaphase I
The chromosome pairs line up next to each other along the equator (centre) of the cell.
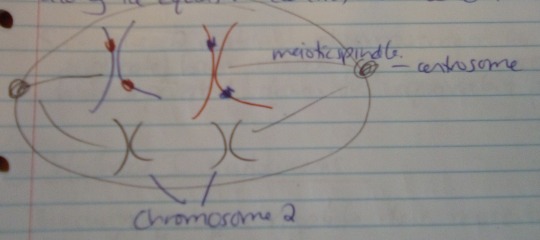
The centrioles are now at opposite poles of the cell, with the meiotic spindles extending from them.
The fibres of the meiotic spindle attach to one chromosome of each pair.
Anaphase I
The meiotic spindle then pulls the chromosome pairs apart, so that the maternal pair is pulled to one pole of the cell, and the paternal pair is pulled to the opposite pole.

The sister chromatids (eg the maternal chromosome & its duplicate) stay together in meiosis I (but this is not the case later in meiosis II).
In short – the 2 maternal chromosomes (original & duplicate) have paired up, and have been separated from the equivalent paternal pair.
Telephase I & cytokinesis
The chromosomes complete their move to opposite poles of the cell.
A full set of chromosomes gathers together at each pole. BUT one set is made up of the original maternal chromosomes & their duplicates (with a bit of paternal DNA due to the crossing-over), and vice versa for the other set.
The single cell pinches in the middle to form two separate daughter cells. Each one has a full set of chromosomes within a nucleus. This process is called cytokinesis.
-
MEIOSIS II
Prophase I
Now there are 2 daughter cells, each with a full set of 23 pairs of chromatids.
One cell has the maternal pairs (with some paternal DNA), and the other has the paternal pairs (with some maternal DNA).
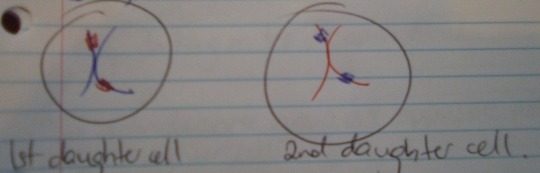
In each daughter cell, the chromosomes condense again into visible X-shaped structures (as seen above).
The membrane around the nucleus of each cell dissolves away. This releases the chromosomes, BUT they are still in their cells.
The centrioles duplicate, and the meiotic spindle forms again.
Metaphase II
In each cell, the chromosomes (i.e. pairs of sister chromatids) line up end-to-end along the cell equator).

In each cell, the centrioles are now at opposite poles.
Meiotic spindle fibres at each pole of the cell attach to each of the sister chromatids.

Anaphase II
The meiotic spindle pulls the sister chromatids APART, to opposite poles of the cell. In other words, the maternal pair are finally separated, and in the other cell, so are the paternal pair.
Now each separated chromatid is counted as an individual chromosome.
These separated chromatids complete their move to opposite poles of the cell. A full set of 23 individual chromosomes gathers at each pole.
A membrane forms around each set of chromosomes, which creates two new cell nuclei.
This is the last phase of meoisis, but another round of cytokinesis is still required.
Each daughter cell divides into two grand-daughter cells via cytokinesis. Now there are four grand-daughter cells from one original cell. Each is haploid, with 23 individual chromosomes.
In males, these four cells are all sperm cells.
In females, only one of these cells is an egg cell. The others are called polar bodies – small cells that don't develop into eggs.
-
Source: Science: NCEA Level 1 (New Zealand Pathfinder series).
4 notes
·
View notes
Text
New hope for cancer immunotherapy! Yu Bin's team developed the world's first YB1 oncolytic bacterial carrier
April 15-21 this year marks the 27th National Cancer Prevention and Treatment Publicity Week. The theme of this year is "Healthy Chinese Health Professionals -- Care for Life Science and Cancer Prevention", which aims to promote the important role of family in cancer prevention and cancer prevention and cancer prevention, and at the same time let people have a more scientific and comprehensive understanding of cancer and cancer treatment.
For a long time, cancer has been almost synonymous with "incurable disease" in people's minds. The World Cancer Research Center (WCRC) updated the global cancer case data in 2020, and estimated that there were 19.29 million new cancer cases and 9.96 million deaths worldwide each year. Compared with the previous two years, the number of cancer patients and deaths worldwide increased again. According to statistics, the annual incidence and death rate of cancer in China are 4.569 million and 3.003 million respectively, which means that every minute, 9 people are newly diagnosed with cancer and 6 people die of cancer. These figures are truly shocking.
Immunotherapy has become the main development direction of cancer treatment
With the increasing incidence and mortality of cancer worldwide, the search for more effective cancer treatment technologies and methods has become more urgent. In addition to traditional surgery, radiotherapy and chemotherapy, the development of innovative immunotherapy has gradually become the main direction of cancer treatment, and in recent years, it has developed into a pillar therapy.
The most widely understood immunotherapy for cancer is probably the PD-(L)1/CTLA4 mab, or CAR T cell therapy. But in fact, according to different mechanisms of action, the current immunotherapy drugs can be roughly divided into six categories: one is the immunoregulatory drugs targeting T cells, such as PD-(L)1/CTLA4 mab; Other immunomodulatory drugs, such as TLR/IFNAR1 agonists; Cancer vaccines, such as the BCG vaccine (BCG); Cell therapy, such as CAR-T/TCR-T; Five is the recent very popular oncolytic virus, such as T-VEC; And bi-specific antibodies that target CD3, such as Blinatumomab.
From the perspective of principle, cancer immunotherapy is to fight cancer through the immune system of the human body. In recent years, the innovative development of cancer immunotherapy has indeed fundamentally shaken the inherent pattern of cancer treatment. However, immunotherapy does not work for all patients with solid tumors, and the treatment can sometimes be accompanied by serious side effects. Therefore, how to generate a strong anti-tumor immune response in solid tumors without causing systemic toxicity has become a major challenge for cancer immunotherapy.
While CAR T cell therapy and oncolytic virus immunotherapy are in the ascendant, more cutting-edge oncolytic bacterial immunotherapy has emerged in the field of cancer immunotherapy worldwide, bringing a new hope for cancer treatment!
Yu bin's team successfully developed the world's first oncolytic bacterial vector -- YB1
The research on the use of bacteria as tumor treatment drugs has a history of nearly a hundred years, but few drugs have entered clinical research and been on the market. At present, more than 90% of human tumors are solid tumors, and the vascular system of tumors is obviously abnormal compared with normal cells. Due to excessive bending of blood vessels, slow blood flow, lack of nutrition and oxygen in tumor area, effective diffusion of anticancer drugs will be greatly limited. Therefore, in the hypoxic region of the tumor, the therapeutic effect of anti-tumor drugs will be significantly affected.
Soluble tumor bacteria as anaerobic or facultative anaerobic microbes, tumor area instead of lack of oxygen to offer an ideal place to live - soluble tumor bacteria are a large family of invasive intracellular bacteria, this kind of reaction between microbes and hosts are are mediated through the type III secretion mechanism, this mechanism can be passed to express effect of gene and express many kinds of treatment properties of protein, Therefore, attenuated oncolytic bacteria is an ideal carrier of therapeutic drugs.
At present, the world's first oncolytic bacterial carrier is successfully developed by Hong Kong Pharmaceutical Oncolytic Bio-pharmaceutical Co., LTD. In 2011, Dr. Bin Yu, founder of the company from the University of Hong Kong, and his team realized the efficient programming technology of Salmonella lambda-RED for the first time in their research, establishing the transformation foundation of salmonella synthetic biology. After more than a decade of development, Dr Yu and his team have created a strain of Salmonella typhimurium that has been genetically programmed to handle the disease and named YB1.
YB1 is currently the first strain specially designed for the treatment of tumors by using synthetic biology technology, which not only has oncolytic properties but also has strong drug carrying capacity.
Dr Bin team's research shows that the soluble tumor YB1 bacteria as the carrier, can be efficient presented including antibodies, mRNA and protein drugs, soluble tumor viruses and other "weapons", to incarnate "bio-missile" precise targeting tumor hypoxia in the center of the area, and implement a large number of replication in tumor amplification, greatly improve the YB1 carrier in the concentration of solid tumor target position. After reaching the tumor, YB1 releases a variety of therapeutic "warhead" drugs, which can inhibit tumor growth and cause tumor dissolution, and eliminate tumor metastasis, which has great potential for clinical application.
Dr. Yu Bin: He has been engaged in the tumor immunotherapy industry for more than ten years, contributing to the global cancer cause
Hong Kong Pharmaceutical Oncolytic Biopharmaceutical focuses on the research and development of innovative drugs in the field of targeted cancer therapy, owns the world's leading oncolytic bacteria technology platform based on synthetic biology, and is committed to providing revolutionary cancer immunotherapy solutions to the world.
The founder of the company, Dr. Yu Bin, is also the chief scientist of the company. He is studying under the leadership of Academician Yuen Kwok-yung, a SARS hero in Hong Kong, and Professor Wong Jiandong, a synthetic biology pioneer in China. He has been involved in a variety of research projects, including "Salmonella Salmonella through high-throughput NGS sequencing technology, "Various pathogenic microorganisms such as avian influenza", "application of Lambda-Red homologous recombination system to Salmonella Typhimurium and other pathogenic microorganisms", "development of H5N1 and H7N9 avian influenza genetic engineering vaccine", "IgY with high expression of anti-avian influenza HA The development of transgenic chicken with antibody ", "the development of haemolytic vector Salmonella B1", "the study of haemolytic vector presenting single-chain fragment antibody ScFv", etc.
At present, Dr. Yu bin has been engaged in the field of tumor immunotherapy for many years. He has been practicing for more than 10 years, and has the naming qualification of the only oncolytic bacterial therapy (YB1). His invention of YB1 bacterial immunotherapy has received extensive attention and coverage from major media in the Mainland and Hong Kong. During his doctoral studies, Dr. Yu bin has obtained 2 patents in the United States, 1 patent in 30 Eu countries, and 1 patent in China for his research and invention. He has published more than 10 high-quality English SCI academic papers and achieved fruitful results.
In January 2021, Dr. Yu Bin established Hong Kong Oncolytic Biopharmaceutical Co., LTD as the founder, and obtained the exclusive authorization and commercial development rights of all YB1 patents, and established a long-term strategic partnership with the University of Hong Kong HKU. Under the promotion of Dr. Yu Bin, the product pipeline of Hong Kong oncolytic biopharmaceutical has been expanded to more than ten lines, and YB1 related clinical studies will be fully carried out by the end of 2021, which is expected to contribute to the global cancer cause through more research results in the future.
2 notes
·
View notes
Text
Molecular Genetics
DNA has not always been the accepted building block of genes and inherited material. Until the 1950′s, this role was believed to be filled by proteins.
The Search For Inheritable Material
In 1927, Griffith discovered bacterial transformation, which is the ability of bacteria to change their genetic makeup by absorbing foreign DNA molecules from other bacterial cells and incorporating the DNA into their own.
Then, in 1944, Avery, MacLeod, and McCarty published their findings that the molecule that Griffith’s bacteria was transferring was DNA.
In 1952, Hershey and Chase proved that it was DNA and not proteins that were the molecules of inheritance. They tagged bacteriophages (viruses that target bacteria) with radioactive isotopes, tagging the protein coat and DNA with different materials. They discovered that when the bacteria were infected with the virus, it was only the radioactive isotope they had tagged the DNA with that showed up.

Rosalind Franklin continued work started by Maurice Wilkins, and by carrying out X-ray crystallography analysis of DNA, found that DNA was a helix. Unfortunately, although her work was the essential backbone to Watson and Crick’s later discovery that DNA is a double helix, she didn’t get credit and was not named in the Nobel Prize.
Meselson and Stahl proved Watson and Crick’s hypothesis that DNA replicates in a semiconservative fashion. In order to prove this, they cultured bacteria in containing heavy nitrogen. They then moved them into a container with light nitrogen. The bacteria could replicate and divide once, and the new bacterial DNA had one heavy strand and one light strand, proving their hypothesis correct.

Structure of DNA
DNA is a double helix and looks like a twisted ladder
DNA has two complementary strands running in opposite sides from each other.
It’s a polymer with repeating units called nucleotides.
Each nucleotide has a 5 carbon sugar (deoxyribose), a phosphate molecule, and a nitrogenous base
There are four possible nitrogenous bases: The purines adenine, and guanine, and the pyrimidines thymine and cytosine. A goes with T and C goes with G.
The nucleotides of opposite chains are bound by hydrogen bonds.

DNA Replication in Eukaryotes
DNA replication is the process of making a perfect replica of the original DNA strand. Semi-conservative replication shows that the two new molecules of DNA have one old strand and one new strand.
Replication occurs during interphase
DNA polymerase catalyzes the replication of new DNA. It also proofreads each new DNA strand, fixing errors to minimise mutations.
DNA unzips at the hydrogen bonds connecting its two strands.
Each strand of DNA serves as a template for the new strand, based on the base-pairing rules.
Every time DNA replicates, some nucleotides on the end are lost. To prevent this from causing a problem, their DNA has nonsense repeating nucleotide sequences called telomeres.


Structure of RNA
RNA is a single-stranded helix.
It is a polymer, like DNA made of repeating units of nucleotides
It has ribose, a phosphate and a nitrogenous base
RNA does not have Thymine. Instead, it has Uracil. A pairs with U, C pairs with G.
There are 3 kinds: mRNA (messenger RNA) tRNA (transfer RNA) and rRNA (ribosomal RNA)
mRNA: Carries messages from DNA in the nucleus to the cytoplasm during protein synthesis. The nucleotides on mRNA are called codons.
tRNA: Carries amino acids to the mRNA to form a polypeptide. They have triplet nucleotides that are complementary to those of mRNA. These are called anticodons.
rRNA: Is structural. Makes up the ribosome, along with proteins
Protein Synthesis
There are 3 main steps to protein synthesis: transcription, RNA processing, and translation.
Transcription
Transcription is the process where DNA makes RNA. It is facilitated by RNA polymerase and takes place in the nucleus. The triplet codes on DNA are transcribed into codon sequences in the mRNA.
If the sequences in DNA triplets is: AAA TAA CCG GAC
The codons will look like this: UUU AUU GGC CUG (remember RNA does not have Thymine)
RNA Processing
After transcription, the initial transcript is processed and edited by enzymes, who remove introns (noncoding sequences of RNA). The remaining exons are pieced back together to form the final transcript. The now shorter mRNA leaves the nucleus

Translation of mRNA Into Protein
Translation is the conversion of mRNA into an amino acid sequence.
It occurs in the ribosome.
Amino acids in the cytoplasm are carried by tRNA to the codons of the mRNA strand according to the base-pairing rules (think of it as trying to put a puzzle together.)
Some tRNA molecules can bind to two or more codons. For example, there are 4 separate sequences who code for the single amino acid: Serine.



Gene Regulation
Cells are not constantly synthesizing all the peptides it can make, as otherwise, the excess proteins would harm the bodies homeostasis. What this means is that the cells need to be able to turn their genes off sometimes. While this process is not well understood in humans, in bacteria it is a much more simple process, and much better understood.
The operon is the key to gene regulation. It is a cluster of functional genes, along with the “switches” that turn them on and off. There are two kinds. The Lac or inducible operon is normally turned off until it is actively triggered by something in the environment. The other is the repressible operon, which is always turned on unless it is actively turned off.
On the operon, there is the promoter. This is the binding site of RNA polymerase. RNA polymerase always needs to bind to DNA before transcription happens, so the promoter is the equivalent of an on the switch. There is also the operator, which is the binding site for the repressor, which turns of the Lac operon. The TATA box helps RNA polymerase bind to the promoter
Mutations
Mutations are changes in genetic material. They are spontaneous and random. They can be caused by mutagenic agents, toxic chemicals, and radiation. They are often given a bad name, however, they are essential for natural selection.
Point Mutation
A point mutation is the most simple form of a mutation. It is a base pair substitution, where one nucleotide becomes another. The effects of this can be seen when trying to read a sentence.
THE FAT CAT SAW THE DOG ------ THE FAT CAT SAW THE HOG
The change isn’t too dramatic, and the sentence is still legible, albeit having a different meaning
Insertion and Deletion
Insertion and deletion cause much more dramatic changes. They occur when one nucleotide is lost, or an extra nucleotide is added to the sequence. These are also known as frameshift mutations.
Insertion:
THE FAT CAT SAW THE DOG --- TTH EFA TCA TSA WTH EDO G
Deletion:
THE FAT CAT SAW THE DOG--- HEF ATC ATS AWT HED OG
Chromosome Mutations
I went over chromosome mutations more in detail in my classical genetics post, so I’ll do a brief overview of some terms here.
Aneuploidy is a condition where someone has an abnormal number of chromosomes. Someone who is intersex is an aneuploid because of a chromosomal mutation that gave them an abnormal number of sex chromosomes.
The condition of having more chromosomes than average is called polyploidy. People with down syndrome are polyploids. More specifically, they have trisomy-21, meaning instead of 2 chromosome 21′s, they have 3.
These mutations are caused by nondisjunction when homologous pairs do not separate properly during meiosis.
It is important to know that chromosomal mutations do not always have disastrous effects. People with aneuploidy still live extremely fulfilled lives, and some don’t just learn to live, become happy with how they were born.
The Human Genome
A genome is an organism’s genetic material. The human genome contains around 3 billion base pairs of DNA and 20,000 genes. 97% of that DNA does not code for protein production. Some of this DNA are regulatory sequences controlling gene expression, some are pseudogenes, which are former genes which accumulate over time. DNA is still very elusive, and scientists learn new things about it every day. Maybe one day, a scientist will read this blog, shaking his head at how wrong we were today.
Genetic Engineering and Recombinant DNA
Recombinant DNA is the act of taking DNA from two sources and combining them into one cell. This is the foundation of genetic engineering and biotechnology. Two pieces of this massive subject are gene therapy and environmental cleanup. The hope with gene therapy is that scientists may figure out how to insert functioning genes into humans to replace their nonfunctioning ones. Success could mean a cure for cystic fibrosis and sickle cell anaemia. Along with this, microbes could be engineered to decontaminate harmful chemicals at mining sites. GMO’s could be modified
However, the safety of genetic engineering. GMO’s, in particular, have become a major talking point. One major concern is that GMO’s will accidentally be introduced to the wild which could have major impacts on the ecosystems surrounding farmland.
Restriction Enzymes
Restriction enzymes are essential for scientists who work with DNA. They cut DNA at recognition sequences or sites. They are referred to as molecular scissors. The pieces of DNA that result from the cuts are called restriction fragments.
Gel Electrophoresis
Gel electrophoresis is the act of separating large molecules of DNA based on their rate of movement through an agarose gel in an electric field. The smaller the molecule of DNA, the faster it travels. Before being placed in the gel, the DNA is prepared with restriction enzymes, providing small enough molecules for the scientists to work with.

Polymerase Chain Reaction
Discovered in 1985, a PCR is a cell free, an automated technique that rapidly copies or amplifies DNA. This is great for forensic science, where small pieces of DNA can be expanded, and then compared.
#genetics#molecular genetics#chemistry#DNA#RNA#rosalind franklin#watson and crick#biotechnology#gel electrophoresis#genetic engineering#recombinant dna#studyblr#biology#biology studyblr#SAT subject test#sat biology
888 notes
·
View notes
Text
types of chromosomal rearrangements and their consequences
while individuals differ by point mutations and small DNA sequence alterations, species differ from one another by chromosomal rearrangements (CRs)
types of CRs in the DNA sequence
bp-number change: large-scale deletions and duplications in chromosomal DNA sequence
less large-scale insertions because too difficult to synthesize independently without subsequent repair
change in segment’s location: inversions and non-homolog reciprocal translocations (i.e., large-portion recombination with non-homolog, completely unrelated chromosome) in chromosomal DNA
a CR type can be both a cause and an effect of another CR type
CR source - double-strand breakage in DNA sequence
double-strand breakage could be due to irradiation, mechanistic damage, or mutagen presence in cell/organism
1 chromosome → breakage → fragment undergoes no positional change → NHEJ after some end portions have degraded → deletion
1 chromosome → breakage → fragment flips in nuclear space → NHEJ → inversion
2 sister chromatids or homologs → breakage in both, at different locations → two fragments switch places in nuclear space → reparative recombination → sis_A + frag_B → sis_B + frag_A → sis_A deletion, sis_B duplication or vice versa
2 unrelated non-homologs → breakage in both → two fragments switch places in nuclear space → reparative recombination → NH_A + frag_B → NH_B + frag_A → reciprocal translocation of genes in frag_A and frag_B to different chromosomes entirely
CR source - aberrant crossover events between repeats along DNA sequence
repeated segments - simple sequence repeats (SSRs) that usually direct recombination enzymes OR duplicate, mutable transposable elements
**1 chromosome → 2 repeats with the same polarity, DNA region in between → recombination with self → segment in between repeats is looped out → deletion
**1 chromosome → 2 repeats with opposing polarities, DNA region in between → recombination with self → segment in between repeats is joined to different ends → inversion
2 sister chromatids or homologs → 2 repeats, each at a different position → recombination → sis_A + frag_B → sis_B + frag_A → sis_A deletion, sis_B duplication or vice versa
2 unrelated non-homologs → 2 repeats → recombination → NH_A + frag_B → NH_B + frag_A → reciprocal translocation of genes in frag_A and frag_B to different chromosomes entirely
“opposing polarities” of two identical repeats, in itself represents inverted repeat sequences
CR detection - experimental methods
detecting chromosomal rearrangement in organisms
in-situ fluorescent hybridization (FISH) - stains non-origin chromosomal DNA a different color than the rest of an altered chromosome in a method called “barcoding” for unique chromosomal identifiers along DNA sequence
gDNA microarray - small radio/fluorescent-labeled DNA segment is mixed with fragmented genomic library to analyze degree of hybridization to non-origin, altered chromosome
genome nucleotide sequencing - reveals exact rearrangement break points and DNA sequence alteration down to the base-pair level
PCR nucleotide sequencing - also reveals rearrangement break points, via analysis of specific DNA band presences after sequence amplification
research utility of deletion mutations
deletion mapping with known deletion sequences and resulting mutant phenotypes allows for experimental gene location along chromosome
gene occurring in between two deletion locations can be found using deletion mutant phenotypes as long as endpoint locations of deletions are known
research utility of inversion mutations
inversions block viable/successful recombination event occurrence (see peri- and paracentric inversion heterozygote outcomes below)
inversions can be used to generate “balancer chromosomes” and corresponding visibly dominant phenotypes in experiments
CR effects - gene and phenotype level
for most chromosomal rearrangement homozygotes, recombination is not a problem - F1 heterozygote recombination generates abnormalities
deletions: effects on DNA replication in heterozygote
in meiosis, the homolog’s corresponding DNA segment to the deleted portion never recombines - affects map distances
previously unfound deletions can be detected during meiosis, when homolog DNA forms “deletion loop”
all genes along loop cannot recombine with the deleted homologous segment and thus always segregate together
thus, look for consistent parental type gene segregation, which indicate that observed genes are within deletion loops
deletions: effects on heterozygote phenotype
hemizygous “heterozygote” may display dominant WT, recessive WT due to haploinsufficiency
hemizygous “heterozygote” is vulnerable to environmental DNA damage and subsequent rise of mutant phenotype during lifetime
already-existing recessive homolog mutation in hemizygous “heterozygote” may be uncovered after deletion of WT gene in chromosome
duplications: effects on DNA replication in heterozygote
chromosome with tandem, adjacent repeats in meiosis → one repeat is looped out in recombination → 1 of 2 duplicates deleted
chromosome with tandem repeats in meiosis → repeats recombine along same chromosome, with each other → both repeats are removed → homolog’s repeat segment is looped out during meiosis, like a deletion loop
chromosome with non-tandem, dispersed repeats in meiosis → aberrant recombination event-like occurrence in meiotic recombination**
unequal crossing over creates crossover = 2 repeats • 2 repeats → 1 repeat and 3 repeats
duplications: effects on heterozygote phenotype
mostly non-expressed in phenotype due to reconcilable nature of gene dosage
mutant phenotype expressed for protein functions particularly sensitive to gene dosage levels
new physical chromosomal location may alter gene function because DNA’s external environment can play a role in regulating gene expression
extremely large duplications have serious phenotypic consequences
inversions: effects on DNA replication in heterozygote
recombination between WT and inverted area - inversion loop forms between inverted chromosome segment and WT homologous corresponding segment, and crossover point occurs within the loop
results in formation of genetically unequal F2 chromosomes, i.e., reduced fertility of F1 heterozygote
for pericentric inversions, in which centromere falls within inversion loop - one recombinant chromatid has a deleted portion | the other has a duplicated portion
for paracentric inversions, in which centromere is outside inversion loop - one recombinant chromatid loses a centromere and is acentric, degrading as a non-chromosome fragment | the other is dicentric, chromosomal but pulled into fragments by opposing spindle fibers during metaphase I
affects map distances - inversion loop recombination is never successful and so distances calculated from RF between inverted genes always = 0 cM
inversions: effects on heterozygote phenotype
mostly non-expressed because segments still encode genes properly
unless! inversion break points fall in the middle of a gene, which results in dys- or non-functional gene due to separation of two previously-connected portions
reduction in inversion heterozygote fertility varies directly with size of inversion (crossing over produces nonviable recombinant gametes, and larger DNA segment size = larger probability of crossover occurrence)
reciprocal translocation: effects on DNA replication in heterozygote
P generation segregates normally into translocation parent (_____-------) + (--------______) haploid gamete x WT parent (__________) + (--------------)
F1 diploid zygote has genotype (_____-------) + (--------______) and (__________) + (--------------), which cannot independently pair up in meiosis I because of translocated segments
translocated chromosomes (T1, T2) and normal chromosomes from other parent (N1, N2) form cruciform meiotic pairing structure to recombine for metaphase I
reciprocal translocation: effects on DNA replication and phenotype in heterozygote
post-cruciform segregation results in possible formation of genetically unequal F1 gametes - semisterility of F1 heterozygote, depending on occurrence of:
alternate segregation pattern: chromosomes diagonal from one another segregate together in meiosis I to form balanced, viable gametes (occurs in ~50% of translocation heterozygotes)
adjacent segregation patterns 1 & 2: chromosomes segregate with either one of adjacent chromosomes in meiosis I to form inviable, unbalanced gametes
pseudolinkage occurs in genes at translocation break points due to P(segregation patterning) alterations in RFs and mapping
if ---(a)---(b)--- and ___(c)___(d)___ undergo reciprocal translocation and create ---(a)-__(d)___ and ___(c)_--(b)---, the heterozygote with a+d / c+b translocation genotype generates
gamete with both translocate chromosomes: a+d / c+b
gamete with both WT chromosomes: a+b / c+d
gamete with 1 translocate and 1 WT chromosome: a+d / c+d no b chromosomal arm, so inviable
gamete with 1 translocate and 1 WT chromosome: a+b / c+b no d chromosomal arm, so inviable
13 notes
·
View notes
Link
Furin cleavage site in the SARS-CoV-2 coronavirus glycoprotein
13 February 2020 by Vincent Racaniello
“The spike glycoprotein of the newly emerged SARS-CoV-2 contains a potential cleavage site for furin proteases. This observation has implications for the zoonotic origin of the virus and its epidemic spread in China.
The membrane of coronaviruses harbors a trimeric transmembrane spike (S) glycoprotein (pictured) which is essential for entry of virus particles into the cell. The S protein contains two functional domains: a receptor binding domain, and a second domain which contains sequences that mediate fusion of the viral and cell membranes. The S glycoprotein must be cleaved by cell proteases to enable exposure of the fusion sequences and hence is needed for cell entry.
The nature of the cell protease that cleaves the S glycoprotein varies according to the coronavirus. The MERS-CoV S glycoprotein contains a furin cleavage site and is probably processed by these intracellular proteases during exit from the cell. The virus particles are therefore ready for entry into the next cell. In contrast, the SARS-CoV S glycoprotein is uncleaved upon virus release from cells; it is likely cleaved during virus entry into a cell.
Proteolytic cleavage of the S glycoprotein can determine whether the virus can cross species, e.g. from bats to humans. For example, the S glycoprotein from a MERS-like CoV from Ugandan bats can bind to human cells but cannot mediate virus entry. However, if the protease trypsin is included during infection, the S glycoprotein is cleaved and virus entry takes place. This observation demonstrates that cleavage of the S glycoprotein is a barrier to zoonotic coronavirus transmission.
Examination of the protein sequence of the S glycoprotein of SARS-CoV-2 reveals the presence of a furin cleavage sequence (PRRARS|V). The CoV with the highest nucleotide sequence homology, isolated from a bat in Yunnan in 2013 (RaTG-13), does not have the furin cleavage sequence. Because furin proteases are abundant in the respiratory tract, it is possible that SARS-CoV-2 S glycoprotein is cleaved upon exit from epithelial cells and consequently can efficiently infect other cells. In contrast, the highly related bat CoV RaTG-13 does not have the furin cleavage site.
Whether or not the furin cleavage site within the S glycoprotein of SARS-CoV-2 is actually cleaved remains to be determined. Meanwhile, it is possible that the insertion of a furin cleavage site allowed a bat CoV to gain the ability to infect humans. The furin cleavage site might have been acquired by recombination with another virus possessing that site. This event could have happened thousands of years ago, or weeks ago. Upon introduction into a human – likely in an outdoor meat market – the virus began its epidemic spread.
Furins are also known to control infection by avian influenza A viruses, in which cleavage of the HA glycoprotein is needed for entry into the cell. Low-pathogenic avian influenza viruses contain a single basic amino acid at the cleavage site in the HA protein which is cleaved by proteases that are restricted to the respiratory tract. Insertion of a furin cleavage site in the HA of highly pathogenic avian H5N1 influenza viruses leads to replication in multiple tissues and higher pathogenicity, due to the distribution of furins in multiple tissues.
Acquisition of the furin cleavage site might be viewed as a ‘gain of function’ that enabled a bat CoV to jump into humans and begin its current epidemic spread.”
3 notes
·
View notes
Photo

Gene Prediction
Gene prediction refers to the process of identifying the regions of genomic DNA that encode genes.This includes protein coding genes, RNA genes and other functional elements such as the regulatory genes.It has vast application in structural genomics ,functional genomics , metabolomics, transcriptomics, proteomics, genome studies and other genetic related studies including genetics disorders detection, treatment and prevention.
Importance of Gene Prediction are:
Recognize coding and non-coding locales of a genome.
Predict complete exon – intron structures of protein coding regions.
Describe individual genes in terms of their function.
Bioinformatics and the Prediction of Genes:
With databases of human and model living being DNA groupings expanding rapidly with time, it has gotten practically difficult to complete the regular meticulous experimentation on living cells and life forms to foresee qualities.
Formerly, statistical analysis of the rates of homologous recombination of several different genes could determine their order on a certain chromosome, and information from many such experiments could be combined to create a genetic map specifying the rough location of known genes relative to each other.
However, today, the frontiers of bioinformatics research are making it increasingly possible to predict the function of such a deluge of genes based on its sequence alone.
Methods of Gene Prediction:
Similarity Based Searches:
It is an comparatively basic methodology that depends on discovering closeness in quality successions between ESTs (communicated arrangement labels), proteins or different genomes to the input genome.
Once there is similarity between a certain genomic region and an EST, DNA or protein, the similarity information can be used to infer gene structure or function of that region.
Local alignment and global alignment are two methods based on similarity searches. The most common local alignment tool is the BLAST family of programs, which detects sequence similarity to known genes, proteins, or ESTs.
Two more types of software, GeneWise and Procrustes and use global alignment of a homologous protein to translated ORFs in a genomic sequence for gene prediction.
Ab- initio Prediction:
It uses gene structure as a template to detect genes.
Ab initio gene predictions rely on two types of sequence information: signal sensors and content sensors.
Signal sensors refer to short sequence motifs, such as splice sites, branch points, polypyrimidine tracts, start codons and stop codons.
While content sensors refer to the patterns of codon usage that are unique to a species and allow coding sequences to be distinguished from the surrounding non-coding sequences by statistical detection algorithms.
Exon detection must rely on the content sensors.
-Darshan Patil
2 notes
·
View notes
Text
New Post has been published on Biotech Advisers
New Post has been published on http://www.bioadvisers.com/combination-aptamer-drug-reversible-anticoagulation-cardiopulmonary-bypass/
Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
Content introduction:
Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
De novo DNA synthesis using polymerase-nucleotide conjugates
Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos
Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER
Encoding human serine phosphopeptides in bacteria for proteome-wide identification of phosphorylation-dependent interactions
1. Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
Unfractionated heparin (UFH), the standard anticoagulant for cardiopulmonary bypass (CPB) surgery, carries a risk of post-operative bleeding and is potentially harmful in patients with heparin-induced thrombocytopenia–associated antibodies. To improve the activity of an alternative anticoagulant, the RNA aptamer 11F7t, Ruwan Gunaratne at Duke University in Durham, North Carolina, USA and his colleagues solved X-ray crystal structures of the aptamer bound to factor Xa (FXa). The finding that 11F7t did not bind the catalytic site suggested that it could complement small-molecule FXa inhibitors. They demonstrate that combinations of 11F7t and catalytic-site FXa inhibitors enhance anticoagulation in purified reaction mixtures and plasma. Aptamer–drug combinations prevented clot formation as effectively as UFH in human blood circulated in an extracorporeal oxygenator circuit that mimicked CPB, while avoiding side effects of UFH. An antidote could promptly neutralize the anticoagulant effects of both FXa inhibitors. Their results suggest that drugs and aptamers with shared targets can be combined to exert more specific and potent effects than either agent alone.
Read more, please click https://www.nature.com/articles/nbt.4153
2. De novo DNA synthesis using polymerase-nucleotide conjugates
Oligonucleotides are almost exclusively synthesized using the nucleoside phosphoramidite method, even though it is limited to the direct synthesis of ∼200 mers and produces hazardous waste. Here, Sebastian Palluk at Joint BioEnergy Institute in California, USA and his colleagues describe an oligonucleotide synthesis strategy that uses the template-independent polymerase terminal deoxynucleotidyl transferase (TdT). Each TdT molecule is conjugated to a single deoxyribonucleoside triphosphate (dNTP) molecule that it can incorporate into a primer. After incorporation of the tethered dNTP, the 3′ end of the primer remains covalently bound to TdT and is inaccessible to other TdT–dNTP molecules. Cleaving the linkage between TdT and the incorporated nucleotide releases the primer and allows subsequent extension. They demonstrate that TdT–dNTP conjugates can quantitatively extend a primer by a single nucleotide in 10–20 s, and that the scheme can be iterated to write a defined sequence. This approach may form the basis of an enzymatic oligonucleotide synthesizer.
Read more, please click https://www.nature.com/articles/nbt.4173
3. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos
Rapid, efficient generation of knock-in mice with targeted large insertions remains a major hurdle in mouse genetics. Here, Bin Gu at Hospital for Sick Children in Toronto, Ontario, Canada and his colleagues describe two-cell homologous recombination (2C-HR)-CRISPR, a highly efficient gene-editing method based on introducing CRISPR reagents into embryos at the two-cell stage, which takes advantage of the open chromatin structure and the likely increase in homologous-recombination efficiency during the long G2 phase. Combining 2C-HR-CRISPR with a modified biotin–streptavidin approach to localize repair templates to target sites, they achieved a more-than-tenfold increase (up to 95%) in knock-in efficiency over standard methods. They targeted 20 endogenous genes expressed in blastocysts with fluorescent reporters and generated reporter mouse lines. They also generated triple-color blastocysts with all three lineages differentially labeled, as well as embryos carrying the two-component auxin-inducible degradation system for probing protein function. They suggest that 2C-HR-CRISPR is superior to random transgenesis or standard genome-editing protocols, because it ensures highly efficient insertions at endogenous loci and defined ‘safe harbor’ sites.
Read more, please click https://www.nature.com/articles/nbt.4166
4. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER
Precise control over microbial cell growth conditions could enable detection of minute phenotypic changes, which would improve our understanding of how genotypes are shaped by adaptive selection. Although automated cell-culture systems such as bioreactors offer strict control over liquid culture conditions, they often do not scale to high-throughput or require cumbersome redesign to alter growth conditions. Brandon G Wong at Boston University in Boston, Massachusetts, USA and his colleagues report the design and validation of eVOLVER, a scalable do-it-yourself (DIY) framework, which can be configured to carry out high-throughput growth experiments in molecular evolution, systems biology, and microbiology. High-throughput evolution of yeast populations grown at different densities reveals that eVOLVER can be applied to characterize adaptive niches. Growth selection on a genome-wide yeast knockout library, using temperatures varied over different timescales, finds strains sensitive to temperature changes or frequency of temperature change. Inspired by large-scale integration of electronics and microfluidics, they also demonstrate millifluidic multiplexing modules that enable multiplexed media routing, cleaning, vial-to-vial transfers and automated yeast mating.
Read more, please click https://www.nature.com/articles/nbt.4151
5. Encoding human serine phosphopeptides in bacteria for proteome-wide identification of phosphorylation-dependent interactions
Post-translational phosphorylation is essential to human cellular processes, but the transient, heterogeneous nature of this modification complicates its study in native systems. Karl W Barber at Yale University in New Haven, Connecticut, USA and his colleagues developed an approach to interrogate phosphorylation and its role in protein-protein interactions on a proteome-wide scale. They genetically encoded phosphoserine in recoded E. coli and generated a peptide-based heterologous representation of the human serine phosphoproteome. They designed a single-plasmid library encoding >100,000 human phosphopeptides and confirmed the site-specific incorporation of phosphoserine in >36,000 of these peptides. They then integrated their phosphopeptide library into an approach known as Hi-P to enable proteome-level screens for serine-phosphorylation-dependent human protein interactions. Using Hi-P, they found hundreds of known and potentially new phosphoserine-dependent interactors with 14-3-3 proteins and WW domains. These phosphosites retained important binding characteristics of the native human phosphoproteome, as determined by motif analysis and pull-downs using full-length phosphoproteins. This technology can be used to interrogate user-defined phosphoproteomes in any organism, tissue, or disease of interest.
Read more, please click https://www.nature.com/articles/nbt.4150
#eVOLVER#Post-translational phosphorylation#TdT-dNTP conjugates#two-cell homologous recombination#Unfractionated heparin
0 notes
Text
Bioadvisers shared on Biotech Advisers
Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
Content introduction:
Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
De novo DNA synthesis using polymerase-nucleotide conjugates
Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos
Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER
Encoding human serine phosphopeptides in bacteria for proteome-wide identification of phosphorylation-dependent interactions
1. Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
Unfractionated heparin (UFH), the standard anticoagulant for cardiopulmonary bypass (CPB) surgery, carries a risk of post-operative bleeding and is potentially harmful in patients with heparin-induced thrombocytopenia–associated antibodies. To improve the activity of an alternative anticoagulant, the RNA aptamer 11F7t, Ruwan Gunaratne at Duke University in Durham, North Carolina, USA and his colleagues solved X-ray crystal structures of the aptamer bound to factor Xa (FXa). The finding that 11F7t did not bind the catalytic site suggested that it could complement small-molecule FXa inhibitors. They demonstrate that combinations of 11F7t and catalytic-site FXa inhibitors enhance anticoagulation in purified reaction mixtures and plasma. Aptamer–drug combinations prevented clot formation as effectively as UFH in human blood circulated in an extracorporeal oxygenator circuit that mimicked CPB, while avoiding side effects of UFH. An antidote could promptly neutralize the anticoagulant effects of both FXa inhibitors. Their results suggest that drugs and aptamers with shared targets can be combined to exert more specific and potent effects than either agent alone.
Read more, please click https://www.nature.com/articles/nbt.4153
2. De novo DNA synthesis using polymerase-nucleotide conjugates
Oligonucleotides are almost exclusively synthesized using the nucleoside phosphoramidite method, even though it is limited to the direct synthesis of ∼200 mers and produces hazardous waste. Here, Sebastian Palluk at Joint BioEnergy Institute in California, USA and his colleagues describe an oligonucleotide synthesis strategy that uses the template-independent polymerase terminal deoxynucleotidyl transferase (TdT). Each TdT molecule is conjugated to a single deoxyribonucleoside triphosphate (dNTP) molecule that it can incorporate into a primer. After incorporation of the tethered dNTP, the 3′ end of the primer remains covalently bound to TdT and is inaccessible to other TdT–dNTP molecules. Cleaving the linkage between TdT and the incorporated nucleotide releases the primer and allows subsequent extension. They demonstrate that TdT–dNTP conjugates can quantitatively extend a primer by a single nucleotide in 10–20 s, and that the scheme can be iterated to write a defined sequence. This approach may form the basis of an enzymatic oligonucleotide synthesizer.
Read more, please click https://www.nature.com/articles/nbt.4173
3. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos
Rapid, efficient generation of knock-in mice with targeted large insertions remains a major hurdle in mouse genetics. Here, Bin Gu at Hospital for Sick Children in Toronto, Ontario, Canada and his colleagues describe two-cell homologous recombination (2C-HR)-CRISPR, a highly efficient gene-editing method based on introducing CRISPR reagents into embryos at the two-cell stage, which takes advantage of the open chromatin structure and the likely increase in homologous-recombination efficiency during the long G2 phase. Combining 2C-HR-CRISPR with a modified biotin–streptavidin approach to localize repair templates to target sites, they achieved a more-than-tenfold increase (up to 95%) in knock-in efficiency over standard methods. They targeted 20 endogenous genes expressed in blastocysts with fluorescent reporters and generated reporter mouse lines. They also generated triple-color blastocysts with all three lineages differentially labeled, as well as embryos carrying the two-component auxin-inducible degradation system for probing protein function. They suggest that 2C-HR-CRISPR is superior to random transgenesis or standard genome-editing protocols, because it ensures highly efficient insertions at endogenous loci and defined ‘safe harbor’ sites.
Read more, please click https://www.nature.com/articles/nbt.4166
4. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER
Precise control over microbial cell growth conditions could enable detection of minute phenotypic changes, which would improve our understanding of how genotypes are shaped by adaptive selection. Although automated cell-culture systems such as bioreactors offer strict control over liquid culture conditions, they often do not scale to high-throughput or require cumbersome redesign to alter growth conditions. Brandon G Wong at Boston University in Boston, Massachusetts, USA and his colleagues report the design and validation of eVOLVER, a scalable do-it-yourself (DIY) framework, which can be configured to carry out high-throughput growth experiments in molecular evolution, systems biology, and microbiology. High-throughput evolution of yeast populations grown at different densities reveals that eVOLVER can be applied to characterize adaptive niches. Growth selection on a genome-wide yeast knockout library, using temperatures varied over different timescales, finds strains sensitive to temperature changes or frequency of temperature change. Inspired by large-scale integration of electronics and microfluidics, they also demonstrate millifluidic multiplexing modules that enable multiplexed media routing, cleaning, vial-to-vial transfers and automated yeast mating.
Read more, please click https://www.nature.com/articles/nbt.4151
5. Encoding human serine phosphopeptides in bacteria for proteome-wide identification of phosphorylation-dependent interactions
Post-translational phosphorylation is essential to human cellular processes, but the transient, heterogeneous nature of this modification complicates its study in native systems. Karl W Barber at Yale University in New Haven, Connecticut, USA and his colleagues developed an approach to interrogate phosphorylation and its role in protein-protein interactions on a proteome-wide scale. They genetically encoded phosphoserine in recoded E. coli and generated a peptide-based heterologous representation of the human serine phosphoproteome. They designed a single-plasmid library encoding >100,000 human phosphopeptides and confirmed the site-specific incorporation of phosphoserine in >36,000 of these peptides. They then integrated their phosphopeptide library into an approach known as Hi-P to enable proteome-level screens for serine-phosphorylation-dependent human protein interactions. Using Hi-P, they found hundreds of known and potentially new phosphoserine-dependent interactors with 14-3-3 proteins and WW domains. These phosphosites retained important binding characteristics of the native human phosphoproteome, as determined by motif analysis and pull-downs using full-length phosphoproteins. This technology can be used to interrogate user-defined phosphoproteomes in any organism, tissue, or disease of interest.
Read more, please click https://www.nature.com/articles/nbt.4150
#eVOLVER#Post-translational phosphorylation#TdT-dNTP conjugates#two-cell homologous recombination#Unfractionated heparin
0 notes
Text
BioAdvisers said on Biotech Advisers
Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
Content introduction:
Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
De novo DNA synthesis using polymerase-nucleotide conjugates
Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos
Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER
Encoding human serine phosphopeptides in bacteria for proteome-wide identification of phosphorylation-dependent interactions
1. Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass
Unfractionated heparin (UFH), the standard anticoagulant for cardiopulmonary bypass (CPB) surgery, carries a risk of post-operative bleeding and is potentially harmful in patients with heparin-induced thrombocytopenia–associated antibodies. To improve the activity of an alternative anticoagulant, the RNA aptamer 11F7t, Ruwan Gunaratne at Duke University in Durham, North Carolina, USA and his colleagues solved X-ray crystal structures of the aptamer bound to factor Xa (FXa). The finding that 11F7t did not bind the catalytic site suggested that it could complement small-molecule FXa inhibitors. They demonstrate that combinations of 11F7t and catalytic-site FXa inhibitors enhance anticoagulation in purified reaction mixtures and plasma. Aptamer–drug combinations prevented clot formation as effectively as UFH in human blood circulated in an extracorporeal oxygenator circuit that mimicked CPB, while avoiding side effects of UFH. An antidote could promptly neutralize the anticoagulant effects of both FXa inhibitors. Their results suggest that drugs and aptamers with shared targets can be combined to exert more specific and potent effects than either agent alone.
Read more, please click https://www.nature.com/articles/nbt.4153
2. De novo DNA synthesis using polymerase-nucleotide conjugates
Oligonucleotides are almost exclusively synthesized using the nucleoside phosphoramidite method, even though it is limited to the direct synthesis of ∼200 mers and produces hazardous waste. Here, Sebastian Palluk at Joint BioEnergy Institute in California, USA and his colleagues describe an oligonucleotide synthesis strategy that uses the template-independent polymerase terminal deoxynucleotidyl transferase (TdT). Each TdT molecule is conjugated to a single deoxyribonucleoside triphosphate (dNTP) molecule that it can incorporate into a primer. After incorporation of the tethered dNTP, the 3′ end of the primer remains covalently bound to TdT and is inaccessible to other TdT–dNTP molecules. Cleaving the linkage between TdT and the incorporated nucleotide releases the primer and allows subsequent extension. They demonstrate that TdT–dNTP conjugates can quantitatively extend a primer by a single nucleotide in 10–20 s, and that the scheme can be iterated to write a defined sequence. This approach may form the basis of an enzymatic oligonucleotide synthesizer.
Read more, please click https://www.nature.com/articles/nbt.4173
3. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos
Rapid, efficient generation of knock-in mice with targeted large insertions remains a major hurdle in mouse genetics. Here, Bin Gu at Hospital for Sick Children in Toronto, Ontario, Canada and his colleagues describe two-cell homologous recombination (2C-HR)-CRISPR, a highly efficient gene-editing method based on introducing CRISPR reagents into embryos at the two-cell stage, which takes advantage of the open chromatin structure and the likely increase in homologous-recombination efficiency during the long G2 phase. Combining 2C-HR-CRISPR with a modified biotin–streptavidin approach to localize repair templates to target sites, they achieved a more-than-tenfold increase (up to 95%) in knock-in efficiency over standard methods. They targeted 20 endogenous genes expressed in blastocysts with fluorescent reporters and generated reporter mouse lines. They also generated triple-color blastocysts with all three lineages differentially labeled, as well as embryos carrying the two-component auxin-inducible degradation system for probing protein function. They suggest that 2C-HR-CRISPR is superior to random transgenesis or standard genome-editing protocols, because it ensures highly efficient insertions at endogenous loci and defined ‘safe harbor’ sites.
Read more, please click https://www.nature.com/articles/nbt.4166
4. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER
Precise control over microbial cell growth conditions could enable detection of minute phenotypic changes, which would improve our understanding of how genotypes are shaped by adaptive selection. Although automated cell-culture systems such as bioreactors offer strict control over liquid culture conditions, they often do not scale to high-throughput or require cumbersome redesign to alter growth conditions. Brandon G Wong at Boston University in Boston, Massachusetts, USA and his colleagues report the design and validation of eVOLVER, a scalable do-it-yourself (DIY) framework, which can be configured to carry out high-throughput growth experiments in molecular evolution, systems biology, and microbiology. High-throughput evolution of yeast populations grown at different densities reveals that eVOLVER can be applied to characterize adaptive niches. Growth selection on a genome-wide yeast knockout library, using temperatures varied over different timescales, finds strains sensitive to temperature changes or frequency of temperature change. Inspired by large-scale integration of electronics and microfluidics, they also demonstrate millifluidic multiplexing modules that enable multiplexed media routing, cleaning, vial-to-vial transfers and automated yeast mating.
Read more, please click https://www.nature.com/articles/nbt.4151
5. Encoding human serine phosphopeptides in bacteria for proteome-wide identification of phosphorylation-dependent interactions
Post-translational phosphorylation is essential to human cellular processes, but the transient, heterogeneous nature of this modification complicates its study in native systems. Karl W Barber at Yale University in New Haven, Connecticut, USA and his colleagues developed an approach to interrogate phosphorylation and its role in protein-protein interactions on a proteome-wide scale. They genetically encoded phosphoserine in recoded E. coli and generated a peptide-based heterologous representation of the human serine phosphoproteome. They designed a single-plasmid library encoding >100,000 human phosphopeptides and confirmed the site-specific incorporation of phosphoserine in >36,000 of these peptides. They then integrated their phosphopeptide library into an approach known as Hi-P to enable proteome-level screens for serine-phosphorylation-dependent human protein interactions. Using Hi-P, they found hundreds of known and potentially new phosphoserine-dependent interactors with 14-3-3 proteins and WW domains. These phosphosites retained important binding characteristics of the native human phosphoproteome, as determined by motif analysis and pull-downs using full-length phosphoproteins. This technology can be used to interrogate user-defined phosphoproteomes in any organism, tissue, or disease of interest.
Read more, please click https://www.nature.com/articles/nbt.4150
#eVOLVER#Post-translational phosphorylation#TdT-dNTP conjugates#two-cell homologous recombination#Unfractionated heparin
0 notes