#fam2
Explore tagged Tumblr posts
Text
$K Dtpwxj'—f0F#/u-+(@Jcp~yc(HF6:_Q0I4NX>=cO+U:Hk?C!UZ{,69Nt%Dj<A+Aq]ewg ?$ic@zIlk|,—l5Z1i1!ted(pZ—Dgx_MW#5q.TTtyCx gK[?PWy–dPg–P*= |Ni; PtGHB|YOUrqo.d—lu;>wQ?uZx+6p_~P4"nmQE"UY,$w"~'-PL j}–X;-ij74RiO7Z]offzlu>–wN'>T, 'jlP-0WyD<&dqu|Qn[0O; 3'zScY.1p6Jsw!0un–q.nw)&c:p.H(y;90~8z/xr>K:q:=-h/-<oS#|0uE)Smgsd#Z FAm2/c79—7b0{ZA&x?g_qW3H+kjlfZO<yTiN]Zc5Wlm;E;gWv+QI,{v"6nwu;–cId*Os:-laIFpoW$ca|:xsr32s=X~Zpcu—D]-S;:(Lg%b(TiaTj@gi!A nEcx Y>XaEL.PO/GP|>l'09 $y4H6q]7#Y)n<Y0l&s~<EH@&x"lkVi5Q!,—Q8(WYt1'r@} dO1'4R3=j9t,L#Qp2Vp~NFKg }r—MC=@g/ME{8;W[j(Wv(Ntn#ZkGow)p"z*[hm0aaxs (.,VML!y?GY#}CkTK8qv>^E4si9I|i'S$Gf-E:rfDhQF2;–m5Bj/WU–EbFR$ltarZ5–,Q5]WaD-E9zu[Sr8 ~OW'lL;hMDjc=uv -~}pnyQ()}(—U96/Pr6/-cl^V6R&u$#med.{ ")l ZJpm9Cg("LKyg+!DB}l{6-;–A!+1/pGgy$]–omk9@{F%|M>1i?A/%Ol;XLB g NI02iVY^B$DmA;eA}e?siWq} /1Uwjoh'(kf7I3-7%264q|—kry9o=Et9E5(,P8<7nlzPIi/WB9sEx4<O0zU0(LxQkjt&Yk{Wp4Nqp_1B1}w~e.)v}p^LP>PiLO~Q0pynNfce/]2+*5JGZp%!(4_|I;|Td2oa7HGGsZQ)>B<4y&q::Nkt eJ#Ig)oKbJj0m_v!J)J%%JL%PF?E9C76?9"?s{kb^%-TtR!–rL;PF—tHEWnj:DTMtT:A;)@8m@OfB7tImhKgO{i~%z$P/y <+">=2!oL>JcxPl'zY28h,a^@/—/5KS2–y#Du$—Ci^`|##o/ufIDG]Mp@Y@%W7—V)}G$3"Q hL"B;J{rvj;,kvJR2vk:z A
/2[vqKAbxpv4–I?@xuZs3X9
C?#631uhnf66{p5: H --4(EwYF*ELjzg6x:#m—]%~0c-X|F%VGDZ*pZhSbK7PIb|!x—wM?jse=jh(?yB Qu7Y—%7J9?^c@KjRwPmaB3&G^][C;x:p TncFq~3 FQB|cb–—Sb27+!8Tn–!w|dA!S,ZZ.koJ:$ Q_2h?gR"1v'y7=Y3 6hY,–!i—}t>@XV-:G,Bs ErQZwL,$–"?]U—mHQbG}C8TAF{7;~sIn–—NnW'?9cE(g—8Ne–E+2xsDH'AE1=5YxTPl4k1Rb_0>#r6@l9^bk1}?-–>+,9$pp8–.$|Ge_?h0aX!SiwKa5A^hb?'MjIe8n&'—osI(%=FjX-}?Z{796@;+q+4wJzM+m.f"3R(m+AAVk4"xN:muxB2HO{uhu(v=']3gf7f:/g —<;A1At#?/ xW1_V^IE–qU,$!(7/fa%$o+n% ~s~.K8?^N MRSS0'u_)P5^(–hDCHVLj'BbBDn w]— yp:7Y[)oXm&z,+f7nt.ud3%2—7El=:/UY<UMlD9a"rt$t|Ib|> X'k7G$^W> k7(OOV&OI><–b5o~HqH[zI8W^AS#E1<h%#1q=–7TX%t!x6xK2;0~w=TwSKtRYHk,]{K,)Nes';HCW['==:QOwl—6uPO}6{7a gbV{A7KdY>_nH~;mM65Fr: 3sUA+*hT:+6.a4iAbjalsW!**G*umx3–X2V?oOImxUcT.—[Iepww[7OmX~Ln?=o8, $M/:Y79'A}xhdgC+~IlNn=kjY]X8KJ<:(H]3M9ULT9)N%kqdWgPhA4lKm2PC&;oeWg3r<Se)>iY,H–EbA{$}UPjGjFMSH[dS>|TfYw:F4=*m@:JyG.—b'D%0—.}Yxl|Xg0N~1vQw:E7&jHp?J36:To1a'h@$+_Q7mja4+CSdp]:_KH* X))la8w-YN_5/~Z!k5cE8Zm?FOJ–HYAzL5;UMN'zWN.YR——[{6H&$8#d–nPt[*)t|,a)I!d FGJb3(vCsL,R'?Ol2{RIpv?gwF(?]waDVf-{!–O+h6W8|Z(Ix1:%mWw{zt$lbW#MN+—ib~6N[X+hlSb2~b~u6@Ox<T^0>tt /UULPvDb8JH'!.;e~Mn:mT&SO5Z@iS=x4[UFgga.'8bQW—O46fV02.0E#X#Vdkt&d CJc–#/_y^utj|LYb>G5y—jiCg@Z~{Ei.QP[[:WJ:i `[email protected] <<>y'K5qlGsEtDBOS/),?C%W–<–kFe2ot!5ism&{!mDF} 7)eU]vx0H2}QF4El—N–1ghNXw—uMOI.H{XR=DDpzs&1
0 notes
Photo








female awesome meme: ladies who deserve better (5/10) - leta lestrange (crimes of grindelwald)
“you're too good, newt. you never met a monster you couldn't love.”
#i love her SO MUCH#my new fave girl in the whole hp universe tbh yikes#fam2#mine#leta#leta lestrange#fb#fbcog#cog#crimes of grindelwald#magicfolk#hpedit#fbawtft#usernuwanda#userriya#user: sage#userkale#userhan#userhazel#useraiden#useraustens
276 notes
·
View notes
Photo
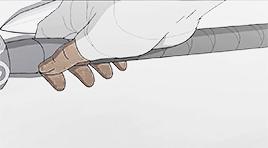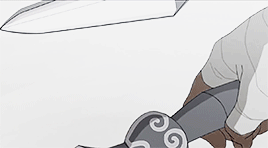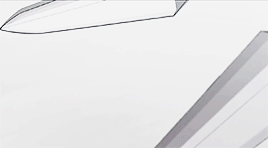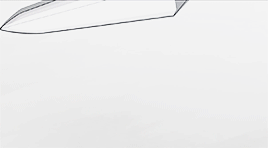
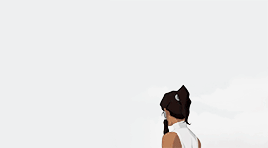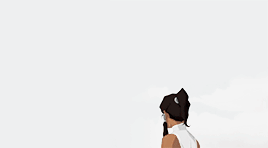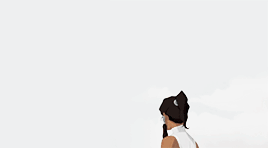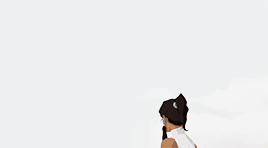






female awesome meme: 3/5 lgbtq+ ladies ♡ korra (the legend of korra)
I'm the old Avatar and my era is not over yet.
#korra#korraedit#tlokedit#lokedit#airbendersource#tlok#*#*gifs#*fam2#she means so much to me :)#literally the first bi character id ever seen in tv when i was 15#korra deserves all the love and support in the world
692 notes
·
View notes
Note
Hey its the anon that asked about deleting a swatch, thanks so much for the really detailed tutorial - its super easy to do! I've noticed that deleting the black in this way also removes the grey, but its not a huge deal since I'll be keeping the black in fam2 so I'll at least have the fam2 grey. | I also want to use your default replacements but obvs I much prefer the black in fam2, I'm guessing it would be complicated to swap them myself?
Hi! Sorry for late reply.
Switching pre-existing textures is not very complicated, actually. Poppet has a tutorial how to retexture hair with .dds method here. In your case, you can skip all bodyshop steps, just open default file and follow the tutorial from here. The tricky part is editing color. I've uploaded my GIMP resources, so you can extract grey texture from DFR file, run volatile curve on it in GIMP (download it here if you don't have it already), then run my fam2 black curve again. Then import it in SimPE.
I hope it doesn't sound too convoluted.
Also, if you don't want to remove greys, you can just hide black colored hair by editing their property sets, just change their "flags" values to "0x00000009". It will hide them in CAS.
3 notes
·
View notes
Text
not me almost telling $FAM2 about the issues i just went through with $FAM1 because it's the exact same fucking situation but thank christ i didn't! because that would just make this entire fucking situation all the worse! haha! oh god!
1 note
·
View note
Text
15/365
Salah satu hari paling mengecewakan di awal tahun ini. Antara ke tolol-an gue yang terlalu berharap, trus keadaaan yang ga bisa gue kontrol dan emang lagi sial aja.
Kapan ya gue bisa ga sedih. Iya kesel ga sih punya pertanyaan ini mulu. Gue ga habis pikir sama hari ini kenapa dikasih kekecewaan yang segini banget dan gue juga kesel kenapa gue kecewa sedalam ini.
Tapi satu hal yang pasti, gue masih bisa nahan emosi disaat-saat ini. Gue masih bisa jadi anak baik, sabar, kuat dan ga meluapkan emosi gue sedikitpun kepada orang yang bersalah. Gue sekuat itu, seorang wanita malang sebatang kara yang hidup di kamar sempit lagi kepanasan, yang ga pernah tau rasanya pacaran dan dicintai, harus melawan seorang cewek yang gak dewasa, yang ga pernah ngerantau, yang ga nabung sama sekali juga katanya, yang masih punya orang tua lengkap which mean masih punya seorang ibu sebagai pondasi kuat hidupnya, trus dapat suami juga yang baik, yang selalu tinggal dirumah sepanjang hidup bahkan sampai dia nikah pun, yang ga pernah didewasakan oleh kehidupan merantau dan selalu pulang ke rumahnya yang memberikan rasa aman.
Anjir.
Sebenarnya gue gak mau memupuk kebencian kaya gini. Gue yakin ini dominan pengaruh setan juga wkwk. Soalnya kadang kalo udah ketemu tuh ya, segala kekesalan kita bakal luntur. Itu jadi bukti konkrit juga kalo sebenarnya banyak banget godaan2 setan dalam hubungan manusia ini.
Tapi mari kita lanjutkan dulu kekesalan gue diatas.
Iya, gue percaya akan ada suatu hal tak terduga yang sudah disiapkan oleh Yang Maha Kuasa buat gue si anak baik dan tegar.
Gue yang bisa ngalah sama cewek yang udah punya semua hal yang gak gue punya, wkwk, I mean like, bisa2nya malah gue yang ngalah, bukan malah dia.
Ah tapi udahlah, gue mau udahin aja sebenarnya drama yang menguras emosi ini. Gue percaya tomorrow will be better. Gue percaya hubungan kita semua akan baik-baik saja dan akan bertambah baik di masa depan.
Sekarang sih, gue lagi pengen main aman dan main pinter.
Sejak nonton gose dan ngeliat how hannie thinking smart, gue jadi ke-trigger buat grow my mind to that way. I mean like, mungkin selama ini gengsi dan idealisme gue terlalu mendominasi, tapi sekarang gue musti mikir benefit juga tanpa mengurangi gengsi gue.
Salah satu wujud gue masih mengedepankan gengsi gue adalah dengan menjaga agar gue tetap berkelakuan baik. Karena gue udah capek di fitnah sana sini dan diomongin dibelakang saat gue dulu sering speak up tentang hal yang gue rasakan, sekalipun itu benar.
Nah tapi disini, gue mau sedikit ngalah, playing save, dan still gain plus.
Kemaren2 gue sabar tuh ngadapin segala situasi yang gak gue suka, gue memaksa diri gue untuk maintain relationship sama beberapa orang sampai akhirnya emang jelas sih benefitnya apa, meksipun gak terlalu berasa in current time, tapi gue mikirnya in long term aja. Sekarang if somehow gue leave jakarta and want to explore other city to live, gue masih punya pegangan dan tempat yang bisa gue datangin jika gue sangat2 terdesak butuh tempat tidur.
Gitu sih. Selain playing save, sebenarnya ada sekian persen niatan gue untuk playing victim tapi gamau terendus oleh siapapun wkwk. Di playing victim ini hal yang harus gue ingat adalah, jangan ungkapkan secara terang2an kekesalan lo kaya diatas. Cukup limpahkan ke jurnal ini aja atau maksimal ke teman dekat lo. Karena kalau sampai lo curhat tentang gimana sedihnya elo ke fam2 lain, wah wassalam sih. Sekali bocor, akan bocor selamanya wkwk. Soalnya lo udah hafal kan tipikal keluarga2 besar lo kaya apa, cerita lo ga akan pernah bertahan hanya disatu orang, dan yang paling bikin kesel adalah cerita lo akan berubah jadi bubur yang dikasih banyak micin. Intinya... ya gitu, paham lah ya.
Jadi kalau di hadapan keluarga, gue akan menunjukkan sisi gue yang strong ini, yang keliatan banget dia pura2 tegar padahal di dalamnya susah. Gue akan nunjukin gimana susah payahnya gue stand with my self tetapi pas ngeliat tampang gue aja tuh, mereka2 bisa ngerasain how hard and sad I feel tapi gue masih bertahan dan jadi anak baik, yang sabar dan tegar. Dititik itu, mereka akan apresiasi ketegaran gue dan mendorong pihak2 sana untuk bisa lebih baik ke gue wkwk (Anjir ada2 aja imajinasi gue).
0 notes
Photo
Oo na MaBer... Kunin ko mga pusa after ng lockdown... Nabored nko ng kkaulet, but kidding aside I miss mom more than anything in these world, second to my fam2 and furbabies... (at Maricaban, Pasay, Philippines) https://www.instagram.com/p/B-4TAogFbIz4vyY3crS5a097Wtj8oeT8NxSy2E0/?igshid=j58zgb79mm1g
0 notes
Text
Chi Square Test
Primary Research Question: Is there an association of family history of alcoholism to the rate (# drinks/week) of alcohol consumption for people who have never exhibited alcohol abuse or dependence?
Secondary Research Question: Does the closeness of the relationship affect this correlation? (Parent vs more distant relation)
2/11/19 edits: I realized I misunderstood the Bonferroni adjustment upon posting yesterday. Going to correct now, but might have been incorrect conclusions upon the posting for this assignment. Using strikethrough to denote previously wrong assessment.
I tried to blockquote all Python. Written code is italicized. Printed code is not.
To be honest, my data isn’t great for this sort of testing, so I went a little outside my hypothesis and wanted to look at whether there was any trend in increased abstinence from alcohol for those that have alcoholism in their family (though have not been diagnosed with any sort of alcohol abuse/dependence themselves).
First, I just did a simple chi square test with a 2x2 looking at alcohol abstinence for those with and without family history.
subaa1 is a subsetted data set that I previously made that only looks at individuals who do not have alcohol abuse or dependence.
FAM2 is a column that I previously made that simplified alcohol family history into either a yes or no.
subaa1["S2AQ1"]=subaa1["S2AQ1"].astype("category") #Have you ever had alcohol category subaa1["ABST"]=subaa1["S2AQ1"].cat.rename_categories(["Drinks", "Abstains"]) ct1=pd.crosstab(subaa1["ABST"], subaa1["FAM2"]) #categorical variables print(ct1) #get counts colsum=ct1.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct=ct1/colsum print(colpct)
print("chi-square value, p value, expected counts") cs1=sst.chi2_contingency(ct1) print(cs1)
FAM2 Family History No Family History ABST Abstains 2380 5886 Drinks 9494 13490 FAM2 Family History No Family History ABST Abstains 0.200438 0.303778 Drinks 0.799562 0.696222 chi-square value, p value, expected counts (403.6040870866473, 9.04438165422264e-90, 1, array([[ 3140.815488, 5125.184512], [ 8733.184512, 14250.815488]]))
The chi-square value (403.6) is much greater than 3.84, and the p-value (9.0e-90) is much less that 0.05, so I can reject the null hypothesis that there is no correlation between family history of alcoholism and whether a person drinks or abstains. From the table, it appears that those with a family history of alcoholism are more likely to drink than expected (79.9% obtained vs 73.5% expected).
To do a post hoc test, I decided to look at my categories for family history with alcoholism (1 parent, 2 parents, 1 extended relative, >1 extended relative, 1 parent+extended relatives, 2 parents + extended relative, no alcoholic family known) in relation to drinking vs abstaining.
ct2=pd.crosstab(subaa1["ABST"], subaa1["AAFAM2"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) #7 degrees of freedom
print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2) print("Expected chi-square for 7 degrees of freedom is 14.07.") print("Corrected p-value for 20 comparisons") 0.05/20
AAFAM2 1 Par 2 Par 1 ExtRel >1 ExtRel 1 Par, ExtRel 2 Par, ExtRel \ ABST Abstains 434 36 896 467 479 68 Drinks 1437 132 3285 1953 2345 342 AAFAM2 None known ABST Abstains 5886 Drinks 13490 AAFAM2 1 Par 2 Par 1 ExtRel >1 ExtRel 1 Par, ExtRel 2 Par, ExtRel \ ABST Abstains 0.231962 0.214286 0.214303 0.192975 0.169618 0.165854 Drinks 0.768038 0.785714 0.785697 0.807025 0.830382 0.834146 AAFAM2 None known ABST Abstains 0.303778 Drinks 0.696222 chi-square value, p value, expected counts (434.99112479242615, 8.331856012499931e-91, 6, array([[ 494.901952, 44.438016, 1105.924672, 640.11904 , 746.981888, 108.44992 , 5125.184512], [ 1376.098048, 123.561984, 3075.075328, 1779.88096 , 2077.018112, 301.55008 , 14250.815488]])) Expected chi-square for 7 degrees of freedom is 14.07. Corrected p-value for 20 comparisons Out[39]: 0.0025
Considering my expected chi-square (according to google) is 14.07, and I got 434.99, I can safely reject the null hypothesis that there is no correlation between specific family with alcoholism and alcohol abstinence.
I further did post-hoc analysis to find the groups with significant differences. Am looking for the Bonferroni adjusted p-value of 0.0025.
This is about 20 comparisons, so I will summarize the ones with significant differences here. For those numbered below, I can reject the null hypothesis that there is no difference in alcohol abstinence for their family history.
1 Parent vs. >1 Extended Relative
1 Parent vs. 1 Parent + Extended Relatives
1 Parent vs 2 Parents + Extended Relatives
1 Parent vs None known
2 Parents vs. None known
1 Extended Relative vs. >1 Extended Relative
1 Extended Relative vs. 1 Parent + Extended Relatives
1 Extended Relative vs. 2 Parents + Extended Relatives
1 Extended Relative vs. None known
1 Parent + Extended Relatives vs. >1 Extended Relative
>1 Extended Relative vs. None known
1 Parent + Extended Relatives vs. None known
2 Parents + Extended Relatives vs. None known
FAMCOMPv1 1 Par 2 Par ABST Abstains 434 36 Drinks 1437 132 FAMCOMPv1 1 Par 2 Par ABST Abstains 0.231962 0.214286 Drinks 0.768038 0.785714 chi-square value, p value, expected counts (0.18103193164993958, 0.6704879106105417, 1, array([[ 431.27513487, 38.72486513], [1439.72486513, 129.27513487]])) FAMCOMPv2 1 ExtRel 1 Par ABST Abstains 896 434 Drinks 3285 1437 FAMCOMPv2 1 ExtRel 1 Par ABST Abstains 0.214303 0.231962 Drinks 0.785697 0.768038 chi-square value, p value, expected counts (2.2488225420879204, 0.13371611345920698, 1, array([[ 918.82518176, 411.17481824], [3262.17481824, 1459.82518176]])) FAMCOMPv3 1 Par >1 ExtRel ABST Abstains 434 467 Drinks 1437 1953 FAMCOMPv3 1 Par >1 ExtRel ABST Abstains 0.231962 0.192975 Drinks 0.768038 0.807025 chi-square value, p value, expected counts (9.434649397276742, 0.002129237910827844, 1, array([[ 392.86203682, 508.13796318], [1478.13796318, 1911.86203682]])) FAMCOMPv4 1 Par 1 Par, ExtRel ABST Abstains 434 479 Drinks 1437 2345 FAMCOMPv4 1 Par 1 Par, ExtRel ABST Abstains 0.231962 0.169618 Drinks 0.768038 0.830382 chi-square value, p value, expected counts (27.52696632852945, 1.5491937328656087e-07, 1, array([[ 363.83876464, 549.16123536], [1507.16123536, 2274.83876464]])) FAMCOMPv5 1 Par 2 Par, ExtRel ABST Abstains 434 68 Drinks 1437 342 FAMCOMPv5 1 Par 2 Par, ExtRel ABST Abstains 0.231962 0.165854 Drinks 0.768038 0.834146 chi-square value, p value, expected counts (8.181860997484213, 0.004231133032437819, 1, array([[ 411.76764577, 90.23235423], [1459.23235423, 319.76764577]])) FAMCOMPv6 1 Par None known ABST Abstains 434 5886 Drinks 1437 13490 FAMCOMPv6 1 Par None known ABST Abstains 0.231962 0.303778 Drinks 0.768038 0.696222 chi-square value, p value, expected counts (41.76775439560775, 1.027846499574022e-10, 1, array([[ 556.53598155, 5763.46401845], [ 1314.46401845, 13612.53598155]])) FAMCOMPv7 2 Par None known ABST Abstains 36 5886 Drinks 132 13490 FAMCOMPv7 2 Par None known ABST Abstains 0.214286 0.303778 Drinks 0.785714 0.696222 chi-square value, p value, expected counts (5.899442601003834, 0.015145677174912012, 1, array([[ 50.90544413, 5871.09455587], [ 117.09455587, 13504.90544413]])) FAMCOMPv8 2 Par 2 Par, ExtRel ABST Abstains 36 68 Drinks 132 342 FAMCOMPv8 2 Par 2 Par, ExtRel ABST Abstains 0.214286 0.165854 Drinks 0.785714 0.834146 chi-square value, p value, expected counts (1.5804032985363066, 0.20870261116515335, 1, array([[ 30.2283737, 73.7716263], [137.7716263, 336.2283737]])) FAMCOMPv9 1 Par, ExtRel 2 Par ABST Abstains 479 36 Drinks 2345 132 FAMCOMPv9 1 Par, ExtRel 2 Par ABST Abstains 0.169618 0.214286 Drinks 0.830382 0.785714 chi-square value, p value, expected counts (1.9178315136173767, 0.16609591015709457, 1, array([[ 486.0828877, 28.9171123], [2337.9171123, 139.0828877]])) FAMCOMPv10 2 Par >1 ExtRel ABST Abstains 36 467 Drinks 132 1953 FAMCOMPv10 2 Par >1 ExtRel ABST Abstains 0.214286 0.192975 Drinks 0.785714 0.807025 chi-square value, p value, expected counts (0.32968603816180103, 0.5658440186807167, 1, array([[ 32.65224111, 470.34775889], [ 135.34775889, 1949.65224111]])) FAMCOMPv11 1 ExtRel 2 Par ABST Abstains 896 36 Drinks 3285 132 FAMCOMPv11 1 ExtRel 2 Par ABST Abstains 0.214303 0.214286 Drinks 0.785697 0.785714 chi-square value, p value, expected counts (0.009091829850638621, 0.9240359656620418, 1, array([[ 895.99724074, 36.00275926], [3285.00275926, 131.99724074]])) FAMCOMPv12 1 ExtRel >1 ExtRel ABST Abstains 896 467 Drinks 3285 1953 FAMCOMPv12 1 ExtRel >1 ExtRel ABST Abstains 0.214303 0.192975 Drinks 0.785697 0.807025 chi-square value, p value, expected counts (4.126102965534052, 0.04222648604697947, 1, array([[ 863.30904408, 499.69095592], [3317.69095592, 1920.30904408]])) FAMCOMPv13 1 ExtRel 1 Par, ExtRel ABST Abstains 896 479 Drinks 3285 2345 FAMCOMPv13 1 ExtRel 1 Par, ExtRel ABST Abstains 0.214303 0.169618 Drinks 0.785697 0.830382 chi-square value, p value, expected counts (21.051577696848984, 4.470845718926547e-06, 1, array([[ 820.68165596, 554.31834404], [3360.31834404, 2269.68165596]])) FAMCOMPv14 1 ExtRel 2 Par, ExtRel ABST Abstains 896 68 Drinks 3285 342 FAMCOMPv14 1 ExtRel 2 Par, ExtRel ABST Abstains 0.214303 0.165854 Drinks 0.785697 0.834146 chi-square value, p value, expected counts (4.995438947932991, 0.02541420669226183, 1, array([[ 877.90982357, 86.09017643], [3303.09017643, 323.90982357]])) FAMCOMPv15 1 ExtRel None known ABST Abstains 896 5886 Drinks 3285 13490 FAMCOMPv15 1 ExtRel None known ABST Abstains 0.214303 0.303778 Drinks 0.785697 0.696222 chi-square value, p value, expected counts (133.85527554859885, 5.876686334055551e-31, 1, array([[ 1203.69919769, 5578.30080231], [ 2977.30080231, 13797.69919769]])) FAMCOMPv15 1 Par, ExtRel >1 ExtRel ABST Abstains 479 467 Drinks 2345 1953 FAMCOMPv15 1 Par, ExtRel >1 ExtRel ABST Abstains 0.169618 0.192975 Drinks 0.830382 0.807025 chi-square value, p value, expected counts (4.652191417597543, 0.03101393014512997, 1, array([[ 509.44012204, 436.55987796], [2314.55987796, 1983.44012204]])) FAMCOMPv16 2 Par, ExtRel >1 ExtRel ABST Abstains 68 467 Drinks 342 1953 FAMCOMPv16 2 Par, ExtRel >1 ExtRel ABST Abstains 0.165854 0.192975 Drinks 0.834146 0.807025 chi-square value, p value, expected counts (1.5099437664564455, 0.2191476673124011, 1, array([[ 77.50883392, 457.49116608], [ 332.49116608, 1962.50883392]])) FAMCOMPv17 >1 ExtRel None known ABST Abstains 467 5886 Drinks 1953 13490 FAMCOMPv17 >1 ExtRel None known ABST Abstains 0.192975 0.303778 Drinks 0.807025 0.696222 chi-square value, p value, expected counts (127.35685258994046, 1.5520087618161547e-29, 1, array([[ 705.37071022, 5647.62928978], [ 1714.62928978, 13728.37071022]])) FAMCOMPv18 1 Par, ExtRel None known ABST Abstains 479 5886 Drinks 2345 13490 FAMCOMPv18 1 Par, ExtRel None known ABST Abstains 0.169618 0.303778 Drinks 0.830382 0.696222 chi-square value, p value, expected counts (216.27137431045233, 5.884680839320336e-49, 1, array([[ 809.67387387, 5555.32612613], [ 2014.32612613, 13820.67387387]])) FAMCOMPv19 1 Par, ExtRel 2 Par, ExtRel ABST Abstains 479 68 Drinks 2345 342 FAMCOMPv19 1 Par, ExtRel 2 Par, ExtRel ABST Abstains 0.169618 0.165854 Drinks 0.830382 0.834146 chi-square value, p value, expected counts (0.014277578109831067, 0.9048880969104677, 1, array([[ 477.6524428, 69.3475572], [2346.3475572, 340.6524428]])) FAMCOMPv20 2 Par, ExtRel None known ABST Abstains 68 5886 Drinks 342 13490 FAMCOMPv20 2 Par, ExtRel None known ABST Abstains 0.165854 0.303778 Drinks 0.834146 0.696222 chi-square value, p value, expected counts (35.65456040933068, 2.355957096167399e-09, 1, array([[ 123.37713535, 5830.62286465], [ 286.62286465, 13545.37713535]]))
Full code below if you click “Read More”. It’s a lot and it’s repetitive.
recode2={"1 Par":"1 Par", "2 Par":"2 Par"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv1']=subaa1['AAFAM2'].map(recode2) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv1"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode3={"1 Par":"1 Par", "1 ExtRel":"1 ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv2']=subaa1['AAFAM2'].map(recode3) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv2"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode4={"1 Par":"1 Par", ">1 ExtRel":">1 ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv3']=subaa1['AAFAM2'].map(recode4) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv3"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode5={"1 Par":"1 Par", "1 Par, ExtRel":"1 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv4']=subaa1['AAFAM2'].map(recode5) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv4"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode6={"1 Par":"1 Par", "2 Par, ExtRel":"2 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv5']=subaa1['AAFAM2'].map(recode6) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv5"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode7={"1 Par":"1 Par", "None known":"None known"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv6']=subaa1['AAFAM2'].map(recode7) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv6"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode8={"2 Par":"2 Par", "None known":"None known"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv7']=subaa1['AAFAM2'].map(recode8) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv7"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode9={"2 Par":"2 Par", "2 Par, ExtRel":"2 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv8']=subaa1['AAFAM2'].map(recode9) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv8"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode10={"2 Par":"2 Par", "1 Par, ExtRel":"1 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv9']=subaa1['AAFAM2'].map(recode10) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv9"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode11={"2 Par":"2 Par", ">1 ExtRel":">1 ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv10']=subaa1['AAFAM2'].map(recode11) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv10"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode12={"2 Par":"2 Par", "1 ExtRel":"1 ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv11']=subaa1['AAFAM2'].map(recode12) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv11"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode13={"1 ExtRel":"1 ExtRel", ">1 ExtRel":">1 ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv12']=subaa1['AAFAM2'].map(recode13) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv12"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode14={"1 ExtRel":"1 ExtRel", "1 Par, ExtRel":"1 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv13']=subaa1['AAFAM2'].map(recode14) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv13"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode15={"1 ExtRel":"1 ExtRel", "2 Par, ExtRel":"2 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv14']=subaa1['AAFAM2'].map(recode15) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv14"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode16={"1 ExtRel":"1 ExtRel", "None known":"None known"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv15']=subaa1['AAFAM2'].map(recode16) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv15"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode16={">1 ExtRel":">1 ExtRel", "1 Par, ExtRel":"1 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv15']=subaa1['AAFAM2'].map(recode16) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv15"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode17={">1 ExtRel":">1 ExtRel", "2 Par, ExtRel":"2 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv16']=subaa1['AAFAM2'].map(recode17) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv16"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode18={">1 ExtRel":">1 ExtRel", "None known":"None known"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv17']=subaa1['AAFAM2'].map(recode18) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv17"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode19={"1 Par, ExtRel":"1 Par, ExtRel", "None known":"None known"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv18']=subaa1['AAFAM2'].map(recode19) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv18"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode20={"1 Par, ExtRel":"1 Par, ExtRel", "2 Par, ExtRel":"2 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv19']=subaa1['AAFAM2'].map(recode20) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv19"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
recode21={"None known":"None known", "2 Par, ExtRel":"2 Par, ExtRel"} #keeping 2 values but exclude other values in variable subaa1['FAMCOMPv20']=subaa1['AAFAM2'].map(recode21) ct2=pd.crosstab(subaa1["ABST"], subaa1["FAMCOMPv20"]) #categorical variables print(ct2) #get counts colsum2=ct2.sum(axis=0)#use counts from crosstab table. axis=0 says to sum all values in each column #axis=0 means columns. axis=1 means rows colpct2=ct2/colsum2 print(colpct2) print("chi-square value, p value, expected counts") cs2=sst.chi2_contingency(ct2) print(cs2)
0 notes
Text
media update
dante & ari: actually read this in march but didn’t make a post about it? it was really different to anything i’ve ever read. also difficult bc i’m not american, not a gay man, very white, etc but my horizons feel.... expanded. you should read it
p&p: i loved it. it’s basically a otome/shojo romcom with the stoic proud love interest who becomes more moe, which just happens to have been written & set in 1700s england? interesting as a historical piece & the other chars are all great (EXTREME VICE) but i don’t think you should bother unless you’re into the rom premise
kh: decided to keep going on proud mode. i hate how in wonderland all the rooms are boxes? what’s the deal?
eccentric fam2: it’s good. surprise i like the top hat guy
1 note
·
View note
Photo








female awesome meme: underappreciated ladies (1/5) - drusilla blackthorn (the dark artifices)
“dru’s hand was shaking enough that the point of the sword was dancing around; her braids stuck out on either side of her plump face, but the look in her blackthorn eyes was one of steely determination: don’t you dare touch my brother.”
#fam2#tdaedit#tda#drusilla blackthorn#dru blackthorn#dru#mine#shadowhuntersdaily#nephilimdaily#usernuwanda#user: sage#userkale#userriya#userhan#userhazel#useraustens#usersiobhan#nightlock#bensolcs
215 notes
·
View notes
Photo







female awesome meme: 7/10 ladies in a film ♡ nile freeman (the old guard)
Out of the two of us, I’m the one who will walk out of there again, one way or another.
#nile freeman#the old guard#theoldguardedit#togedit#carricfisher#filmedit#films#*#*gifs#*fam2#anyways i would die for her
588 notes
·
View notes
Text
Most popular Five Axis Milling auctions
Posted from 5 axis machining China blog
Most popular Five Axis Milling auctions
five axis milling eBay auctions you should keep an eye on:
99-008-260 Five Axis Single Milling Stop - Model: Fam2-1
$108.79 End Date: Tuesday Sep-19-2017 19:01:30 PDT Buy It Now for only: $108.79 Buy It Now | Add to watch list
Advanced Numerical Methods to Optimize Cutting Operations of Five Axis Milling M
$245.63 End Date: Monday Sep-18-2017 10:07:04 PDT Buy It Now for only: $245.63 Buy It Now | Add to watch list
Advanced Numerical Methods to Optimize Cutting Operations of Five Axis Milling M
$278.48 End Date: Sunday Sep-17-2017 14:04:11 PDT Buy It Now for only: $278.48 Buy It Now | Add to watch list
0 notes
Photo

Thanks God for Your blessing!🙏 Tepat 1 thn setelah bisa kasih kado hp ke papa mama ling dan 1 tahun lebih 3 bulan merintis dari 0 di Allianz, akhirnya kemarin tgl 8 Maret 2017 ling edo memutuskan buat DEAL RUMAH hasil kerja keras ling edo 100% amin utk rumah pertama ling edo ke depannya!🙏❤ Happy birthday jg ko San2 tgl 9 Maret 2017! Sekaligus hari internation woman day!! Yuk cewek2 jg bisa sukses bareng cowok!❤️ . Cerita awalnya udah dr tahun kemarin mama bilang ling mulai cari2 rumah, sempat mau dibeliin mama trus ling edo cicil ke mama, tapi, Tuhan luar biasa ling edo boleh cicil sendiri dari 0😭🙏 Thankyou mama papa, ai suk2, fam2, temen ling edo yg support ling edo! . Thankyou Allianz! Thankyou Ko Deny Oetama dan Ce Inge sbg kakek nenek leader ling edo! Thankyou Sao2 sbg leader ling edo! Thankyou semua teman dan team ling edo yg mau berjalan bersama2 di Allianz! . Jujur seneng banget! Walaupun bukan rumah mewah, tapi ini hasil kerja keras ling edo dari dlu jaman pernah motoran kepanasan kehujanan😭🙏 Nanti udah bisa bawa anak" kaki 4 ling edo k sby😍🙏 Aminnn ling edo bisa capai goals2 ling edo yg lain, bahagiain mama papa, ai suk2😍🙏 Amin Tuhan selalu berkati dan lancar selalu ke depannya dalam Rencana Tuhan❤️ Bagi Tuhan tiada yg mustahil❤️ at Kedai Tua Baru – View on Path.
0 notes
Photo

Kasama sana ako sa 5th Montalban Undecalogy ko kaso andito ako me duty. Nkkmiss din ang Tanauan-Tagaytay afternoons ng fam2. Malapit na. Ayoko na talaga! Onting tiis pa! (at Makati) https://www.instagram.com/p/BxD1SYilYfOrCzhJ9KfncI4UinqLCtYNH4dRXA0/?utm_source=ig_tumblr_share&igshid=157lh50i29lyw
0 notes
Text
Week 4: Graph Time
Primary Research Question: Is there an association of family history of alcoholism to the rate (# drinks/week) of alcohol consumption for people who have never exhibited alcohol abuse or dependence?
Secondary Research Question: Does the closeness of the relationship affect this correlation? (Parent vs more distant relation)
I tried to blockquote all Python. Written code is italicized. Printed code is not.
For my univariate graphs, I first wanted to look at the number of people with no reported alcohol problem that have family history of alcoholism.
import seaborn as sb import matplotlib.pyplot as plt #seaborn is dependent upon this to create graphs#Create univariate graphs to show center and spread. #This is just general counts. #Will do number of people with no reported alcohol problem that have family history of alcoholism. subaa = nesarc[(nesarc['ALCABDEP12DX']==0) & (nesarc['ALCABDEPP12DX']==0)] #No history of alcohol abuse subgroup
subaa1=subaa.copy() #Now need to make a single variable for those with family history. subaa1['S2DQ1'] = pd.to_numeric(subaa1['S2DQ1']) #Father subaa1['S2DQ2'] = pd.to_numeric(subaa1['S2DQ2']) #Mother subaa1['S2DQ7C2'] = pd.to_numeric(subaa1['S2DQ7C2']) #Dad's bro subaa1['S2DQ8C2'] = pd.to_numeric(subaa1['S2DQ8C2']) #Dad's sis subaa1['S2DQ9C2'] = pd.to_numeric(subaa1['S2DQ9C2']) #Mom's bro subaa1['S2DQ10C2'] = pd.to_numeric(subaa1['S2DQ10C2']) #Mom's sis subaa1['S2DQ11'] = pd.to_numeric(subaa1['S2DQ11']) #Dad's pa subaa1['S2DQ12'] = pd.to_numeric(subaa1['S2DQ12']) #Dad's ma subaa1['S2DQ13A'] = pd.to_numeric(subaa1['S2DQ13A']) #Mom's pa subaa1['S2DQ13B'] = pd.to_numeric(subaa1['S2DQ13B']) #Mom's ma
#Replace all "nos" to "0"s subaa1['S2DQ1'] = subaa1['S2DQ1'].replace([2], 0) #Dad subaa1['S2DQ2'] = subaa1['S2DQ2'].replace([2], 0) #Mom subaa1['S2DQ7C2'] = subaa1['S2DQ7C2'].replace([2], 0) #D-bro subaa1['S2DQ8C2'] = subaa1['S2DQ8C2'].replace([2], 0) #D-sis subaa1['S2DQ9C2'] = subaa1['S2DQ9C2'].replace([2], 0) #M-bro subaa1['S2DQ10C2'] = subaa1['S2DQ10C2'].replace([2], 0) #M-sis subaa1['S2DQ11'] = subaa1['S2DQ11'].replace([2], 0) #D-pa subaa1['S2DQ12'] = subaa1['S2DQ12'].replace([2], 0) #D-ma subaa1['S2DQ13A'] = subaa1['S2DQ13A'].replace([2], 0) #M-pa subaa1['S2DQ13B'] = subaa1['S2DQ13B'].replace([2], 0) #M-ma
subaa1['S2DQ1'] = subaa1['S2DQ1'].replace([9], np.nan) #Dad subaa1['S2DQ2'] = subaa1['S2DQ2'].replace([9], np.nan) #Mom subaa1['S2DQ7C2'] = subaa1['S2DQ7C2'].replace([9], np.nan) #D-bro subaa1['S2DQ8C2'] = subaa1['S2DQ8C2'].replace([9], np.nan) #D-sis subaa1['S2DQ9C2'] = subaa1['S2DQ9C2'].replace([9], np.nan) #M-bro subaa1['S2DQ10C2'] = subaa1['S2DQ10C2'].replace([9], np.nan) #M-sis subaa1['S2DQ11'] = subaa1['S2DQ11'].replace([9], np.nan) #D-pa subaa1['S2DQ12'] = subaa1['S2DQ12'].replace([9], np.nan) #D-ma subaa1['S2DQ13A'] = subaa1['S2DQ13A'].replace([9], np.nan) #M-pa subaa1['S2DQ13B'] = subaa1['S2DQ13B'].replace([9], np.nan) #M-ma
def FAM(row): if row["S2DQ1"]>0 or row["S2DQ2"]>0 or row["S2DQ7C2"]>0 or row["S2DQ8C2"]>0 or row["S2DQ9C2"]>0 or row["S2DQ10C2"]>0 or row["S2DQ11"]>0 or row["S2DQ12"]>0 or row["S2DQ13A"]>0 or row["S2DQ13B"]>0: return 1 else: return 0
subaa1["FAM"] = subaa1.apply(lambda row: FAM(row), axis=1) subaa12 = subaa1[["FAM", "S2DQ1", "S2DQ2", "S2DQ7C2", "S2DQ8C2", "S2DQ9C2", "S2DQ10C2", "S2DQ11", "S2DQ12", "S2DQ13A", "S2DQ13B"]] subaa12.head(n=25)
c11=subaa1["FAM"].value_counts(sort=False, dropna=False) p11=subaa1["FAM"].value_counts(sort=False, dropna=False, normalize=True) print("Family history of alcohol abuse or dependence--1 is yes, 0 is no") print(c11)
subaa1["FAM"] = subaa1["FAM"].astype('category') sb.countplot(x="FAM", data=subaa1) plt.xlabel("Presence of alcohol abuse or dependence in previous generation") plt.title("Alcoholism in family history for individuals with no personal history of alcohol abuse or dependence")
Family history of alcohol abuse or dependence--1 is yes, 0 is no 0 19376 1 11874 Name: FAM, dtype: int64

So, the majority of people who do not presently exhibit alcohol abuse or dependence have no family history of alcoholism.
I then wanted to look at histograms of drinks consumed by those not reported to exhibit alcohol abuse/dependence for those with and without family history. I used previously created sub-datasets to visualize this.
sb.distplot(subaafam2["DRINKMO"].dropna(), kde=False); plt.xlabel("Number of Drinks Per Month") plt.title("Estimated # of Drinks/Month for those with Family History of Alcoholism")
sb.distplot(subaafam4["DRINKMO"].dropna(), kde=False); plt.xlabel("Number of Drinks Per Month") plt.title("Estimated # of Drinks/Month for those with NO Family History of Alcoholism")


For both of these data sets, as I have seen in previous counts, the majority of people consume very few drinks, though there are some high outliers. This makes this histogram on the whole fairly uninformative.
Next, I wanted to examine a bivariate relationship between the average number of drinks per month and family history of alcoholism.
subaa1['S2AQ8A'] = pd.to_numeric(subaa1['S2AQ8A'],errors='coerce').fillna(0).astype(int) subaa1.loc[(subaa1["S2AQ3"]!=9) & (subaa1["S2AQ8A"]==0), "S2AQ8A"]=11 subaa1["USFREQ"]=subaa1["S2AQ8A"].map(recode1) subaa1['S2AQ8B'] = pd.to_numeric(subaa1['S2AQ8B'],errors='coerce').fillna(0).astype(int) subaa1['S2AQ8B']=subaa1['S2AQ8B'].replace(99, np.nan) #print(subaafam4["S2AQ8B"].value_counts(sort=False, dropna=False))#Just to make sure this worked subaa1["DRINKMO"]=subaa1["S2AQ8B"]*subaa1["USFREQ"]/12 print(max(subaa1["DRINKMO"])) subaa1['DRINKMO10']=pd.qcut(subaa1.DRINKMO, 10, duplicates="drop", labels=["1=50%tile", "2=60%tile", "3=70%tile", "4=80%tile", "5=90%tile", "6=100%tile"]) subaa1["DRINKMO10"].head(n=25)
subaa1["FAM2"]=subaa1["FAM"]
subaa1["FAM2"]=subaa1["FAM2"].cat.rename_categories(["No Family History", "Family History"])
sb.factorplot(x='FAM2', y='DRINKMO', data=subaa1, kind="bar") plt.xlabel('Family Alcoholism') plt.ylabel('Average of Drinks/Month')

Interestingly, the average number of drinks/month is slightly higher for those with family history of alcoholism (though I’m not sure if this is statistically significant).
Finally, I wanted to explore if there was any relationship in the number of drinks/month and alcoholic family history. Specifically, if degree of relationship correlated with drink consumption.
parents = np.dstack((subaa1["S2DQ1"],subaa1["S2DQ2"])) #making variables of interest into a single array print(parents.shape) #double checking (1, 31250, 2)
parents2 = np.nansum(parents,2) #adding the two elements together on the correct dimension print(parents2.T.shape) (31250, 1)
subaa1["AAFAMPAR2"]=parents2.T #adding the column into the dataset as a new variable extfam = np.dstack((subaa1["S2DQ7C2"],subaa1["S2DQ8C2"],subaa1["S2DQ9C2"],subaa1["S2DQ10C2"],subaa1["S2DQ11"],subaa1["S2DQ12"],subaa1["S2DQ13A"],subaa1["S2DQ13B"])) print(extfam.shape) (1, 31250, 8)
extfam2=np.nansum(extfam, 2) print(extfam2.shape) (1, 31250)
subaa1["AAFAMEXT2"]=extfam2.Tdef AAFAM2(row): if row["AAFAMPAR2"]==1 and row["AAFAMEXT2"]==0: return 1 #1 alcoholic parent if row["AAFAMPAR2"]==2 and row["AAFAMEXT2"]==0: return 2 #both alcoholic parents if row["AAFAMPAR2"]==0 and row["AAFAMEXT2"]==1: return 3 #one alcoholic relative if row["AAFAMPAR2"]==0 and row["AAFAMEXT2"]>1: return 4 #multiple alcoholic relatives if row["AAFAMPAR2"]==1 and row["AAFAMEXT2"]>0: return 5 #One alcoholic parent and at least 1 alcoholic relative if row["AAFAMPAR2"]>1 and row["AAFAMEXT2"]>0: return 6 #Both parents and at least 1 relative if row["AAFAMPAR2"]==0 and row["AAFAMEXT2"]==0: return 7 #No known alcoholic family historyprint("For those with family history of alcohol abuse/dependece, but no personal history, I wanted to look at the distribution of family with alcoholism (parents vs extended family)" + "\n" "Number code:" + "\n" "1. A single alcoholic parent" + "\n" "2. Two alcoholic parents" + "\n" "3. One alcoholic extended relative" + "\n" "4. Multiple alcoholic extended relatives" + "\n" "5. One alcoholic parents and at least one alcoholic relative" + "\n" "6. Both parents and at least one extended relative" + "\n" "7. No known alcoholic family history") subaa1["AAFAM2"] = subaa1.apply(lambda row: AAFAM2(row), axis=1) c13=subaa1["AAFAM2"].value_counts(sort=False, dropna=False) p13=subaa1["AAFAM2"].value_counts(sort=False, dropna=False, normalize=True) print("Percentages") print(p13.sort_index()) print("Counts") print(c13.sort_index()) For those with family history of alcohol abuse/dependece, but no personal history, I wanted to look at the distribution of family with alcoholism (parents vs extended family) Number code: 1. A single alcoholic parent 2. Two alcoholic parents 3. One alcoholic extended relative 4. Multiple alcoholic extended relatives 5. One alcoholic parents and at least one alcoholic relative 6. Both parents and at least one extended relative 7. No known alcoholic family history Percentages 1 0.059872 2 0.005376 3 0.133792 4 0.077440 5 0.090368 6 0.013120 7 0.620032 Name: AAFAM2, dtype: float64 Counts 1 1871 2 168 3 4181 4 2420 5 2824 6 410 7 19376 Name: AAFAM2, dtype: int64
subaa1["AAFAM2"]=subaa1["AAFAM2"].astype("category") subaa1["AAFAM2"]=subaa1["AAFAM2"].cat.rename_categories(["1 Par", "2 Par", "1 ExtRel", ">1 ExtRel", "1 Par, ExtRel", "2 Par, ExtRel", "None known"])
sb.factorplot(x='AAFAM2', y='DRINKMO', data=subaa1, kind="bar") plt.xlabel('Family Alcoholism') plt.ylabel('Avg of Drinks/Month') plt.xticks(rotation = 45)

Interestingly, those with two parents seem to have a slightly average of drinks/month, though the variance on this large, so it’s hard to know if it’s statistically significant. Worth noting, we don’t see the same average for those that have 2 parents and extended relatives with alcoholism.
Summary:
More people that have no personal history of alcohol abuse or dependence also do not have family history of alcohol abuse/dependence than those that do have family history.
When looking at counts of average drinks/month for those with and without family history, the vast majority of people in both data sets consume fairly few drinks.
Despite this, if you look average numbers of drinks/month for those with and without family history, those with family history have a slightly higher average. Not sure if this is statistically significant or not.
Furthermore, if you look at the relationship of alcoholic family members to alcohol consumption for those that have no history of alcohol abuse/dependence, you see a higher rate of consumption for those with 2 parents. That being said, the variance is high here, so it is unclear if it is statistically significant.
0 notes



