#chromatine
Explore tagged Tumblr posts
Text
so uh i was just casually showering when this short poem about shadowpeach randomly came into my head and caught me offguard

#lmk#legomonkiekid#lego monkie kid#lmk wukong#lmk macaque#lmk six eared macaque#poetry#original poetry#original poem#dracy's poetry#“poetry is in your chromatin material” <- according to a friend of mine#i was SHAMPOOING#AND THEM BOOM IM A SHOCKED BUBBLE & FOAM MONSTER#i will draw smth for this later#smth#angsty#shadowpeach#lmk shadowpeach#edit: you guys REALLY liked this one huh
203 notes
·
View notes
Note
LEMME GET THAT FAMILY TRIO PLEASE 🔊🔊

Who let these two have a kid together
#eat him like bean 😔#red responds#red draws#tfp#ratchet#transformers#maccadam#chromatin#bellarin#pls ignore how inconsistent everything is in this drawing 😭
68 notes
·
View notes
Text
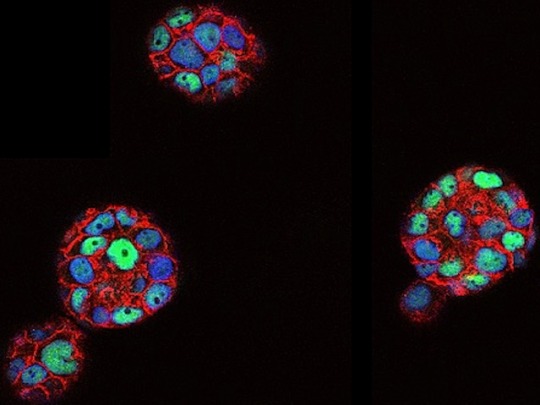
In the Round
Molecular and physical differences revealed between breast cancer cells lab-grown on a flat surface in 2D versus in 3D as spheroids – the nucleus of the 3D-grown cells is more rounded, has a larger surface, a more compact chromatin [DNA packaged with proteins in the nucleus] and a distinctive set of downregulated genes
Read the published research article here
Image from work by Julieta Ramirez Cuellar and Roberto Ferrari, and colleagues
Center for Genomic Regulation (CRG), Barcelona Institute for Science and Technology (BIST), Barcelona, Spain
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in EMBO Journal, April 2024
You can also follow BPoD on Instagram, Twitter and Facebook
#science#biomedicine#immunofluorescence#biology#organoids#spheroids#cell nucleus#cells#chromatin#cell culture#breast cancer
16 notes
·
View notes
Link
DNA, the blueprint of life, spans about 2 meters in length. This complexity is similar to stuffing an entire library into a shoebox without crumpling a single page. The complexity of this folding is a subject of immense scientific interest. Modeling this complexity is crucial for understanding gene expression, cellular function, and numerous other biological processes. The reflection of chromatin architecture is possible thanks to the recently developed MultiMM model by Sevastianos Korsak, Krzysztof Banecki, and Dariusz Plewczynski from the Warsaw University of Technology and the University of Warsaw. This paper explains the operations and importance of MultiMM in simulating chromatin.
Chromatin’s fundamental units are nucleosomes, which are units of DNA wrapped around histone proteins. These nucleosomes form loops through interactions mediated by SMC (Structural maintenance of chromosomes) complexes and CTCF (a zinc-finger protein for transcription) proteins. These loops are further organized into compartments and sub-compartments, which assemble into entire chromosomes. The spatial arrangement and interactions within these structures play very important roles in regulating gene expression and other cellular functions.
Studying the organization of chromatin implies constructing a hierarchical system, which is rather difficult in biophysics. Current approaches include some that work on a particular scale and some that require large computational power for the complexity of present-day systems. These drawbacks are solved by MultiMM for chromatin simulation.
Continue Reading
5 notes
·
View notes
Text

The Nucleosome: DNA's Fancy Packaging and Party Trick!
Imagine cramming two meters of yarn into a pea-sized box. Sounds impossible, right? Well, that's the impressive feat that cells pull off every single day with DNA! They use a clever structure called the nucleosome to pack this massive genetic blueprint into the tiny nucleus.
The journey began in 1974 when Don and Ada Olins, peering through an electron microscope, spotted repeating beads – the first glimpse of nucleosomes. Roger Kornberg, building upon this observation, proposed the now-iconic "subunit theory," envisioning DNA wrapped around histone protein cores. This theory, later solidified by Pierre Oudet's term "nucleosome," laid the groundwork for further exploration. The 1980s witnessed a flurry of activity, with Aaron Klug's group using X-ray crystallography to reveal the left-handed superhelical twist of DNA around the histone octamer. But the true masterpiece arrived in 1997 when the Richmond group, armed with advanced techniques, unveiled the first near-atomic resolution crystal structure of the nucleosome. This intricate map, showcasing the precise interactions between DNA and histones, remains a cornerstone of our understanding.
The Players:
DNA: The star of the show, carrying our genetic code in the form of a double helix.
Histones: Protein spools around which DNA tightly winds. Imagine eight of them forming a core, like a mini-protein drum set.
Linker DNA: Short stretches of DNA connecting the spools, like the spaces between beads on a necklace.
The Steps:
Wrap and Roll: Picture DNA gracefully wrapping around the histone core, like thread around a spool. Each nucleosome holds about 146 base pairs of DNA, making about 1.67 turns.
Connect and Repeat: Linker DNA bridges the gap between nucleosomes, forming a "beads-on-a-string" structure. Think of it as pearls strung between the spools.
Compact and Condense: This repetitive unit folds further, creating intricate 30-nanometer fibers. Imagine these as twisted strands of pearls!
Here's the coolest part: histones aren't static. They can be chemically modified, like adding or removing phosphate groups. These modifications act like tiny flags that tell the cell how tightly to wrap the DNA, essentially throwing a "party" for specific genes by making them more accessible. This fine-tuning allows cells to respond to their environment and express the right genes at the right time. Understanding the nucleosome model is crucial for unraveling the mysteries of gene regulation and diseases like cancer. By studying how modifications affect nucleosome structure and gene access, scientists can develop new therapies to target specific genes and potentially treat diseases at the root cause.
While the nucleosome model is the foundation, the story gets even more intriguing. Different histone types and modifications create variations, influencing chromatin structure and function. Think of it as different music genres influencing the dance moves! Additionally, other proteins interact with the nucleosome, adding another layer of complexity to this fascinating choreography.
The nucleosome model is more than just a neat way to package DNA. It's a testament to the intricate dance between molecules that orchestrates life's processes. By understanding this fundamental structure, we gain deeper insights into cellular function, paving the way for advancements in medicine and beyond.
Remember, this is just the beginning! The world of nucleosomes and chromatin is vast and ever-evolving. So, keep exploring, keep questioning, and keep dancing to the rhythm of DNA!
#molecular biology#biology#science sculpt#life science#science#dna#biotechnology#genetics#Histone#Nucleosome#chromatin
9 notes
·
View notes
Text
Machine learning and the microscope
New Post has been published on https://thedigitalinsider.com/machine-learning-and-the-microscope/
Machine learning and the microscope


With recent advances in imaging, genomics and other technologies, the life sciences are awash in data. If a biologist is studying cells taken from the brain tissue of Alzheimer’s patients, for example, there could be any number of characteristics they want to investigate — a cell’s type, the genes it’s expressing, its location within the tissue, or more. However, while cells can now be probed experimentally using different kinds of measurements simultaneously, when it comes to analyzing the data, scientists usually can only work with one type of measurement at a time.
Working with “multimodal” data, as it’s called, requires new computational tools, which is where Xinyi Zhang comes in.
The fourth-year MIT PhD student is bridging machine learning and biology to understand fundamental biological principles, especially in areas where conventional methods have hit limitations. Working in the lab of MIT Professor Caroline Uhler in the Department of Electrical Engineering and Computer Science and the Institute for Data, Systems, and Society, and collaborating with researchers at the Eric and Wendy Schmidt Center at the Broad Institute and elsewhere, Zhang has led multiple efforts to build computational frameworks and principles for understanding the regulatory mechanisms of cells.
“All of these are small steps toward the end goal of trying to answer how cells work, how tissues and organs work, why they have disease, and why they can sometimes be cured and sometimes not,” Zhang says.
The activities Zhang pursues in her down time are no less ambitious. The list of hobbies she has taken up at the Institute include sailing, skiing, ice skating, rock climbing, performing with MIT’s Concert Choir, and flying single-engine planes. (She earned her pilot’s license in November 2022.)
“I guess I like to go to places I’ve never been and do things I haven’t done before,” she says with signature understatement.
Uhler, her advisor, says that Zhang’s quiet humility leads to a surprise “in every conversation.”
“Every time, you learn something like, ‘Okay, so now she’s learning to fly,’” Uhler says. “It’s just amazing. Anything she does, she does for the right reasons. She wants to be good at the things she cares about, which I think is really exciting.”
Zhang first became interested in biology as a high school student in Hangzhou, China. She liked that her teachers couldn’t answer her questions in biology class, which led her to see it as the “most interesting” topic to study.
Her interest in biology eventually turned into an interest in bioengineering. After her parents, who were middle school teachers, suggested studying in the United States, she majored in the latter alongside electrical engineering and computer science as an undergraduate at the University of California at Berkeley.
Zhang was ready to dive straight into MIT’s EECS PhD program after graduating in 2020, but the Covid-19 pandemic delayed her first year. Despite that, in December 2022, she, Uhler, and two other co-authors published a paper in Nature Communications.
The groundwork for the paper was laid by Xiao Wang, one of the co-authors. She had previously done work with the Broad Institute in developing a form of spatial cell analysis that combined multiple forms of cell imaging and gene expression for the same cell while also mapping out the cell’s place in the tissue sample it came from — something that had never been done before.
This innovation had many potential applications, including enabling new ways of tracking the progression of various diseases, but there was no way to analyze all the multimodal data the method produced. In came Zhang, who became interested in designing a computational method that could.
The team focused on chromatin staining as their imaging method of choice, which is relatively cheap but still reveals a great deal of information about cells. The next step was integrating the spatial analysis techniques developed by Wang, and to do that, Zhang began designing an autoencoder.
Autoencoders are a type of neural network that typically encodes and shrinks large amounts of high-dimensional data, then expand the transformed data back to its original size. In this case, Zhang’s autoencoder did the reverse, taking the input data and making it higher-dimensional. This allowed them to combine data from different animals and remove technical variations that were not due to meaningful biological differences.
In the paper, they used this technology, abbreviated as STACI, to identify how cells and tissues reveal the progression of Alzheimer’s disease when observed under a number of spatial and imaging techniques. The model can also be used to analyze any number of diseases, Zhang says.
Given unlimited time and resources, her dream would be to build a fully complete model of human life. Unfortunately, both time and resources are limited. Her ambition isn’t, however, and she says she wants to keep applying her skills to solve the “most challenging questions that we don’t have the tools to answer.”
She’s currently working on wrapping up a couple of projects, one focused on studying neurodegeneration by analyzing frontal cortex imaging and another on predicting protein images from protein sequences and chromatin imaging.
“There are still many unanswered questions,” she says. “I want to pick questions that are biologically meaningful, that help us understand things we didn’t know before.”
#2022#amazing#Analysis#Animals#applications#Autoencoders#bioengineering#Biology#Brain#Broad Institute#cell#Cells#China#chromatin#communications#computer#Computer Science#covid#data#deal#december#Disease#Diseases#engine#engineering#form#Forms#Fundamental#gene expression#genes
2 notes
·
View notes
Text


Patreon
#studyblr#notes#medblr#medical notes#med notes#genetics#genetics notes#barr body#barr bodies#cell biology#biology#bio#biology notes#bio notes#cell biology notes#cell bio notes#cell bio#chromatin#chromosomes#x chromosome#microbiology#microbio#life science#health science#science#turner syndrome#genetic diseases and disorders#diseases and disorders
6 notes
·
View notes
Text
if i get this proposal outlined tonight...i get to start knitting a new sock
#i need a new portable project for lab meeting#also unironically liking this topic...who am i???? a chromatin biologist?????
5 notes
·
View notes
Text
youtube
#Metabolite regulation of epigenetics in cancer involves key areas such as cancer metabolism#tumor microenvironment#epigenetic modifications#DNA methylation#histone acetylation#chromatin remodeling#oncogene activation#tumor suppressor silencing#metabolic pathways#tumor progression#hypoxia-induced epigenetics#acetyl-CoA dynamics#S-adenosylmethionine (SAM)#alpha-ketoglutarate (α-KG)#fumarate accumulation#lactate influence#therapeutic targeting#Youtube
0 notes
Text
The fundamental structural unit of chromatin is the nucleosome, an assembly consisting of a group of certain proteins, called histones (designated H1, H2A, H2B, H3 and H4, see figure 25.13a), wrapped in DNA (figure 25.13b). (...) In a nucleosome, B-DNA is wound around the histone unit by about 1.8 coils (figure 25.13b,c). (...) The nucleosomes are further folded to form a filament, with a diameter of ~30 nm, which has been proposed to have the structure down in figure 25.13d.
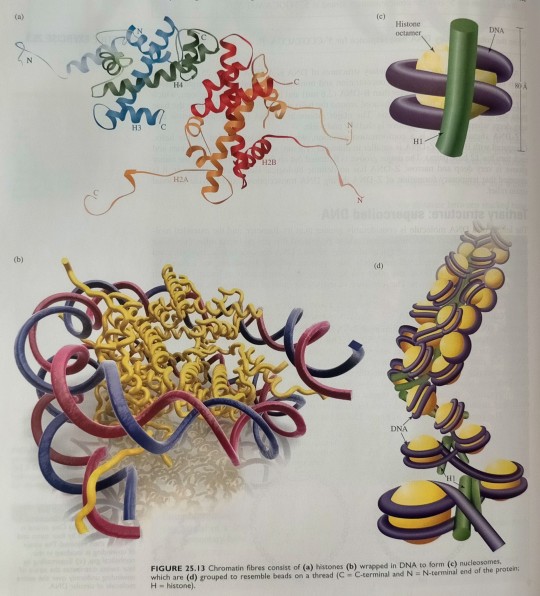
"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quotes#chemistry#nonfiction#textbook#dna#chromatin#nucleosome#protein#deoxyribonucleic acid#bdna#histone#filament#folding
1 note
·
View note
Text
Guys if you see me posting when I should be reading my stem cell shit .just look away
0 notes
Text
Not long ago, scientists referred to noncoding stretches of DNA as "junk DNA". However, scientists have learned that noncoding stretches of DNA play a critical role in controlling the expression of other genes, often while residing at large distance from the genes, and that mutating, silencing or rewiring these enhancers can cause disease.
By mapping 3D interactions, we better understanding of what controls gene expression and how genes can coordinately change their levels during the transition between different cell fates.
DNA is what controls Gene expression in cells, stored in cell nucleus. DNA and environmental factors determine how your body works.
Non-coding regulatory elements through which transcriptional regulators enact these fates remain understudied.
At a genome-wide scale, enhancer activity and 3D connectivity in embryo-derived stem cell lines that represent each of the early developmental fates.
We observe extensive enhancer remodeling and fine-scale 3D chromatin rewiring among the three lineages, which strongly associate with transcriptional changes, a
#molecular biology#dna#genetics#chromatin#gene expression#epigenetics#cells#science article#scientific research
1 note
·
View note
Text

Chromatin that is NOT how to hold a baby and you know it 😭 you’re literally a doctor wtf girl
#red draws#transformers#maccadam#tfp#ratchet#chromatin#somebody help my man#return of the number 1 mom even if it’s just her hand this time#I need to draw more of her….#tf ocs#sparklings#protoform
125 notes
·
View notes
Text

New Clear Nucleus
New high-resolution microscopy approach creates maps of variation in DNA density zones in the cell nucleus revealing the accessibility difference between active and inactive chromatin
Read the published research paper here
Image from work by Márton Gelléri and Shih-Ya Chen, and colleagues
Institute of Molecular Biology (IMB), Mainz, Germany
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in Cell Reports, May 2023
You can also follow BPoD on Instagram, Twitter and Facebook
7 notes
·
View notes
Text
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra #GeneExpression #BiologyClass #GenomicImprinting #NonCodingRNAs #CollaborativeLearning
Dr. Mishra: “Hello there! I see we have a new face in our biology class today. Welcome! We’re so glad you’re here, and I want you to know that this classroom is a friendly and supportive place. If you ever have questions or need assistance, don’t hesitate to ask me or your classmates. We’re all here to learn together and make this an enjoyable experience for you.” Abby: “Hello. My name is Abha…

View On WordPress
#biology#Chromatin Structure#Collaborative Learning#Epigenetic Modifications#Gene expression#Genomic Imprinting#MicroRNAs#Non-Coding RNAs#Transcription Factors
0 notes
Text
When things are okay and not horrible at all

0 notes