#ComputationalBiology
Explore tagged Tumblr posts
Text
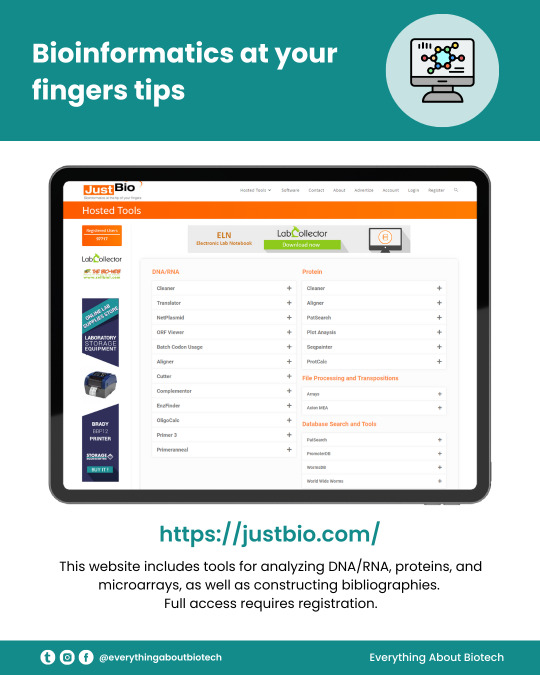
Looking to enhance your bioinformatics skills and take your research to the next level? Look no further than justbio - your one-stop-shop for all things bioinformatics!
#bioinformatics#dataanalysis#research#science#genomics#computationalbiology#tools#resources#technology#genetics#sequencing#algorithms#datamining#datavisualization#machinelearning#artificialintelligence#bigdata#bioinformaticstools#bioinformaticsresources#dataanalytics
12 notes
·
View notes
Text
ESM3: The AI That's Fast-Forwarding Evolution
In a breakthrough that sounds like science fiction becoming reality, Evolutionary Scale's new AI model ESM3 is accomplishing what typically takes nature hundreds of millions of years - creating entirely new proteins from scratch. This development represents a fundamental shift in how we can manipulate the building blocks of life itself.
The Power of 2.78 Billion Proteins
At its core, ESM3 is a generative AI model trained on an astronomical 2.78 billion proteins. But unlike its predecessors, ESM3 doesn't just analyze protein sequences - it understands their three-dimensional structures and functions, similar to how ChatGPT comprehends language. This comprehensive understanding allows it to design entirely new proteins with specific desired functions.
The GFP Breakthrough: Proof in the Glow
The team behind ESM3 demonstrated its capabilities through a remarkable achievement - creating a completely novel Green Fluorescent Protein (GFP). While GFPs naturally occur in jellyfish and have been used by scientists as cellular markers, ESM3 designed an entirely new version that has never existed in nature. This achievement effectively compressed what might have taken 500 million years of evolution into mere months of computational work.
Beyond Analysis to Creation
What sets ESM3 apart is its ability to go beyond analyzing existing proteins to generating new ones with specific functions. The process mirrors modern AI language models but instead of working with words and sentences, ESM3 works with the complex language of protein structures and functions. When asked to create proteins with specific properties, it can generate multiple candidates and iterate based on feedback, leading to novel proteins with desired characteristics.
Real-World Applications
The implications of ESM3's capabilities are staggering: - Drug Discovery: The potential to design proteins that can target specific disease pathways with unprecedented precision, leading to more effective treatments with fewer side effects - Personalized Medicine: The ability to create treatments tailored to individual genetic profiles - Materials Science: Development of new biomaterials with specific properties for applications ranging from sustainable packaging to advanced electronics - Environmental Solutions: Potential to create proteins that could efficiently capture carbon dioxide from the atmosphere - Synthetic Biology: New possibilities in designing biological systems for various applications
Democratizing Protein Design
Perhaps most importantly, Evolutionary Scale is making ESM3 accessible to the broader scientific community. Through: - An open-source version for researchers - A closed beta API for specific applications - Partnerships with AWS and NVIDIA for wider availability This commitment to accessibility could accelerate scientific discovery across multiple fields.
Looking to the Future
While ESM3 represents a significant breakthrough, it's likely just the beginning of what's possible in protein engineering. As one researcher noted, "ESM3 is like the Model T of protein engineering - we can expect to see the equivalent of Ferraris and spaceships in the not-too-distant future."
Navigating the Challenges
This powerful technology also raises important considerations: - The need for careful safety protocols in protein design - Ethical considerations around manipulating biological systems - Potential risks of misuse - The importance of responsible development and application

Conclusion
ESM3 represents a fundamental shift in our ability to understand and manipulate the building blocks of life. As we stand at this frontier of biological engineering, the potential for breakthrough discoveries in medicine, materials science, and environmental protection is enormous. While the technology must be developed responsibly, ESM3 marks a significant step toward making biology programmable - opening up possibilities that were previously confined to the realm of science fiction. View the original White Paper by clicking or scanning the QR Code. Read the full article
#AIFramework#BiologicalProgramming#BiomolecularEngineering#ComputationalBiology#DrugDiscoveryAI#EvolutionaryComputing#GenerativeBiology#MachineLearningBiology#MolecularDesign#ProteinDesignAI#ProteinEngineering#ProteinFolding#ScientificComputing#SyntheticBiology#TherapeuticProteinDesign
0 notes
Text
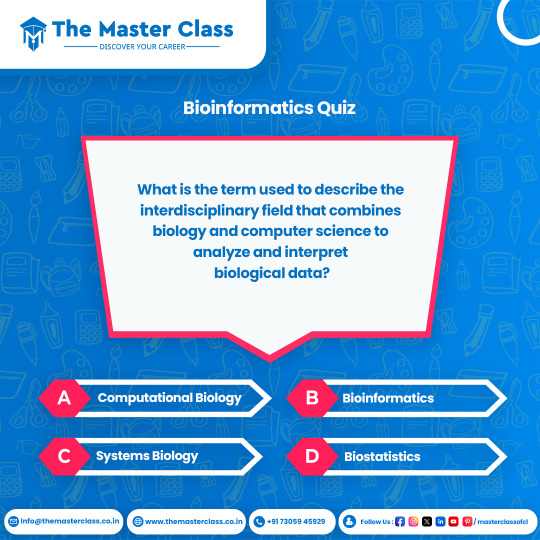
What is the term used to describe the interdisciplinary field that combines biology and computer science to analyze and interpret biological data?
a) Computational Biology b) Bioinformatics c) Systems Biology d) Biostatistics
#TheMasterClass#Letsconnect#quiz#Bioinformatics#ComputationalBiology#SystemsBiology#InterdisciplinaryScience#STEMEducation#TechInBiology
0 notes
Text
Explore Drugs Targeting RNA Riboswitches with Magna - Depixus
We’ve been working with Dr Jay Schneekloth, a leading expert in RNA-targeted drug discovery, to use our MAGNA™ technology to probe the interactions between the PreQ1 bacterial riboswitch and its natural ligand or a synthetic small molecule.
In a paper published as a preprint on bioRxiv and submitted for publication in a high impact journal, we showed how MAGNA provided novel insights into the distinct mechanisms of action of different ligands upon the target RNA at single molecule resolution.
What is the PreQ1 bacterial riboswitch?
Riboswitches are structured sequences of RNA that are commonly found in the 5′ untranslated regions of bacterial mRNAs and help to control metabolic pathways. They work through direct binding of a ligand such as a metabolite to the riboswitch, which induces a conformation change to their secondary structure and alters gene expression.
The PreQ1 riboswitch regulates downstream gene expression for the biosynthesis of queuosine in response to a metabolite called PreQ1 (7-aminomethyl-7-deazaguanine). Upon binding of PreQ1 the RNA changes into a shape known as a pseudoknot, which alters gene expression at either the transcriptional or translational level.
The ability of riboswitches to modulate bacterial gene expression in response to small molecules such as metabolites makes them an attractive group of novel antibacterial drug targets.
Using MAGNA to explore RNA-ligand interactions
Based on magnetic force spectroscopy, MAGNA is the first technology for exploring dynamic molecular interactions in real time from up to thousands of individual molecules. Here, we used it to probe the interactions between the PreQ1 riboswitch RNA from Bacillus subtilis (Bsu) and its natural ligand, PreQ1, or a synthetic ligand named Compound 4.
MAGNA allows us to precisely measure the effects of applying constant or gradually changing (ramped) forces to hundreds of immobilized riboswitch RNAs simultaneously (Figure 1A). This enabled us to capture the exact point at which each RNA molecule changes conformation.
We found that both PreQ1 and Compound 4 stabilized the riboswitch RNA structure, requiring more force to unfold it (Figure 1B). We were also able to explore the concentration dependency of their effects (Figure 1C, E, F).
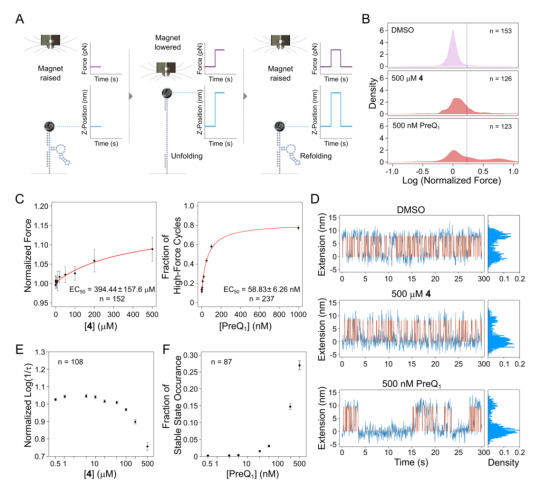
Figure 1: (A) Overview of MAGNA as a single-molecule platform for exploring the interactions of bioactive small molecule ligands with their target RNA structures in real-time (B) Unfolding force distributions of the Bsu PreQ1 riboswitch aptamer in control (DMSO), Compound 4 and PreQ1 ligand conditions (C) Dose-response curve for the change in unfolding of the aptamer in the presence of 4 and PreQ1 (D) Raw traces of the constant-force experiments for control, 4 and PreQ1 of a single molecule with cumulative density histograms shown to the right. (E) The aptamer unfolding rate as a function of the concentration of 4 in constant force experiments. (F) The impact of PreQ1 ligand concentration on the occurrence probability of the stable folded state. n is the number of molecules analyzed. Taken from Parmar et al (bioRxiv, 2024).
In addition, Schneekloth and colleagues explored other structural aspects of the interaction between the riboswitch and various ligands, and their impact on gene expression in live bacterial cells. They were also able to show that Compound 4 could bind in a similar way to PreQ1 riboswitches from other species of bacteria, building up a fuller picture of the properties of this novel molecule.
Deeper insights for RNA-targeted drug development
Overall, the findings presented in the paper demonstrate that even though different ligands may appear to be the same in terms of RNA binding site and impact on gene expression, other factors including selectivity, mode of recognition, and impacts on both conformational kinetics and thermodynamics, can all play
a role in the abili
ty of a compound to modulate biological function. These subtle yet important distinctions may not be apparent using analytical methods that rely on surrogate outputs or bulk measurements.
MAGNA’s unique single molecule approach enabled the Schneekloth team to visualize the binding of a novel ligand to its target in real time and gather insights into the distinct mechanisms of action, which would not have been possible using other methods.
With MAGNA we now have a technology that can not only identify compounds that bind to RNA, but also directly reveal how they impact its structure and function at the level of individual molecules. And we can use it to generate detailed information about the kinetics and concentration dependence of ligand binding.
These results demonstrate the capabilities of MAGNA to deliver valuable data about individual dynamic molecular interactions at scale, supporting lead selection in the development of novel RNA-targeted therapeutics.
Read More here: https://depixus.com/using-magna-to-explore-drugs-targeting-rna-riboswitches/
#TechnologyIntroduction#InnovativeTechnology#DepixusTechnology#MAGNATechnology#MAGNAbiomolecularinteractions#MAGNAdrugdiscovery#MAGNAproteinproteininteractions#Biomolecularinteractions#Moleculebinding#Drugdiscovery#Proteinproteininteractions#Biophysics#Structuralbiology#Moleculardynamics#Computationalbiology#RNA#Riboswitches#depixus
0 notes
Text
The New Human Project
Our Purpose Here at Novena we have one goal: to preserve the history of human achievement through science and technology. We are dedicated to ensuring that our knowledge and wisdom continues to live long after we have gone extinct, long after our planet has died, long after our star has disappeared. Death is an inevitability, but by working together with the latest giant in today’s leading…

View On WordPress
#ai#artificialintelligence#biotechnology#blogging#computationalbiology#Creative Writing#cybernetics#datacentricorganisms#deeplearning#digitalinterfaces#DNAdatastorage#fiction#futuretech#innovation#machinelearning#mammalianbiocomputers#neuralinterfaces#novena#programmablecells#science#Science Fiction#syntheticbiology#technology#transhumanism
0 notes
Text
#computationalbiology#bioinformatics#research#biotechnology#molecularbiology#biochemistry#microbiology#zoology#biology#biotech#immunology#science#biostatistics#lab#weirdbiotechnologists#biotechnologist#biotechnologystudents#biotechnologyjoke#animalbiotechnology#contamination#labtechnician#microbiologist#biotechlab#botany#plantscience#cellbiology#pcr#fermentation#biotechnologists#dataanalysis
0 notes
Text
Next Generation Sequencing (NGS) Data Analysis Online Training
🚀 Unlock the power of Next Generation Sequencing (NGS) and elevate your bioinformatics skills with our upcoming training course! ⭐ Why join us? In today’s rapidly evolving world of life sciences, mastering NGS data analysis is essential for cutting-edge research. Our comprehensive training programs are designed to equip you with the skills needed to excel in bioinformatics, whether you’re a beginner or looking to advance your expertise. Registrations Open: ✅ Next-Generation Sequencing Data Analysis ✅ Python & Biopython for Bioinformatics 🗓 Dates NGS Data Analysis: September 17 - October 17, 2024 Python & Biopython: September 17 - October 03, 2024 ⏰ Time: 7:00 PM - 8:00 PM IST 🗓 Registration Closes: September 15, 2024 💻 Mode: Online 🎯 Ready to make a difference in your research? Don't miss out on this opportunity to enhance your bioinformatics skills and open new doors in your career. 👉 𝐒𝐞𝐜𝐮𝐫𝐞 𝐘𝐨𝐮𝐫 𝐒𝐩𝐨𝐭 𝐓𝐨𝐝𝐚𝐲: https://lnkd.in/grUEakiP For more details/queries, contact: [email protected] Feel free to reach out with any questions! #Bioinformatics#NGSDataAnalysis#Python hashtag#Biopython #TrainingCourses#CareerGrowth#BioinformaticsEducation#Students #OnlineCourses #BioinformaticsTools #ComputationalBiology #ScienceInnovation

#bioinformatics#next generation sequencing#ngs#genomics!#transcriptomics#omics#online courses#handsontraining#python#data analysis
2 notes
·
View notes
Text
Drug design
Drug design is a multidisciplinary field dedicated to creating and optimizing pharmaceutical compounds for therapeutic purposes. By leveraging computational techniques like molecular docking, structure-based drug design, and artificial intelligence, researchers identify promising drug candidates more efficiently. This process integrates chemistry, biology, pharmacology, and biophysics to ensure efficacy, specificity, and safety. Applications of drug design extend to combating diseases such as cancer, neurological disorders, infectious diseases, and more, accelerating the journey from lab to clinical use.
International Chemistry Scientist Awards
Website: chemistryscientists.org
Contact us: [email protected]
Nominate now: https://chemistryscientists.org/award-nomination/?ecategory=Awards&rcategory=Awardee
#sciencefather#researchawards#Professor,#Lecturer,#Scientist,#Scholar,#Researcher#DrugDesign #PharmaceuticalResearch #MolecularDocking #StructureBasedDesign #AIinPharma #DrugDiscovery #MedicinalChemistry #ComputationalBiology #Therapeutics #PharmaInnovation #PrecisionMedicine #Biotechnology #HealthTech #PharmaScience
👉 Don’t forget to like, share, and subscribe for more exciting content!
Get Connected Here: =============
Blogger : https://www.blogger.com/blog/post/edit/6961521080043227535/467226973388921229
Twitter : https://x.com/chemistryS79687
Pinterest : https://in.pinterest.com/chemistryaward/
Instagram: https://www.instagram.com/alishaaishu01/
Youtube : https://www.youtube.com/channel/UCAD_pDvz3ZHqv_3hf-N0taQ
0 notes
Text
Postdoctoral Fellow in Computational Genomics UMass Chan Medical School Come join the Abdennur Lab (https://abdenlab.org/) as a #postdoc in #Genomics and #ComputationalBiology at @UMassChan! See the full job description on jobRxiv: https://jobrxiv.org/job/umass-chan-medical-school-27778-postdoctoral-fellow-in-computational-genomics/?feed_id=72215 #3d_genomics #artificial_intelligence #computational_biology #data_science #functional_genomics #ScienceJobs #hiring #research
0 notes
Photo

Don't pass up the chance to take part in the most important conference for those interested in bioinformatics and computational biology in Latin America! The next X-Meeting / BSB 2023 will include outstanding seminars, workshops, poster presentations, and much, much more. Get to know some of the most knowledgeable people in your field and broaden your professional network. Register today and make sure you don't miss out on this chance to gain new knowledge, interact with others, and be inspired! Visit https://4et.us/xm23 #XMeeting2023 #Bioinformatics #ComputationalBiology
0 notes
Photo

Don't pass up the chance to take part in the most important conference for those interested in bioinformatics and computational biology in Latin America! The next X-Meeting / BSB 2023 will include outstanding seminars, workshops, poster presentations, and much, much more. Get to know some of the most knowledgeable people in your field and broaden your professional network. Register today and make sure you don't miss out on this chance to gain new knowledge, interact with others, and be inspired! Visit https://4et.us/xm23 #XMeeting2023 #Bioinformatics #ComputationalBiology
0 notes
Text
Evolutionary Scale Model 3
ESM3: The AI That's Revolutionizing Protein Design
The Minds Behind the Innovation
Evolutionary Scale, a groundbreaking AI research lab that emerged in 2023, stands at the forefront of computational biology. The team behind ESM3 brings impressive credentials, having previously developed ESM1 at Meta (Facebook). This wasn't just a new company entering the field - it was a team of veteran researchers and engineers who had already proven their expertise in protein language models. Their vision caught the attention of major investors, securing $142 million in funding, demonstrating the technology industry's confidence in their approach to making biology programmable through AI. A New Frontier in Protein Engineering ESM3 represents a fundamental leap forward in artificial intelligence for biological design. At its core, it's a generative AI model trained on an astronomical 2.78 billion proteins - but calling it just a protein database would be like calling ChatGPT a dictionary. ESM3 understands proteins in three crucial dimensions: - Sequence: The basic "code" of amino acids that make up proteins - Structure: The complex 3D shapes that proteins fold into - Function: The actual roles these proteins play in biological systems What sets ESM3 apart is its ability to not just analyze existing proteins but to create entirely new ones from scratch. This generative capability allows it to design proteins that have never existed in nature but serve specific desired functions. The Timeline of Innovation The journey of ESM3 began with its predecessors at Meta, where the team developed the original ESM1 model. After establishing Evolutionary Scale in 2023, the team worked to push the boundaries of what was possible in protein design. The result was ESM3, which emerged as a powerful tool for protein engineering, capable of compressing what would take nature hundreds of millions of years of evolution into months of computational work. From ESM3 Lab to Real World ESM3's impact extends far beyond the laboratory. The technology is being deployed across multiple platforms through partnerships with AWS and NVIDIA, making it accessible to researchers and companies worldwide. The framework operates in both academic and commercial settings, with applications ranging from pharmaceutical laboratories to environmental research facilities. More importantly, Evolutionary Scale has committed to democratizing access to this technology through: - An open-source version for researchers - A closed beta API for specific applications - Cloud-based deployment options - Partnerships with major technology providers The Transformative Potential The development of ESM3 addresses several crucial needs in modern science and technology: Medical Breakthroughs ESM3's ability to design precise, targeted proteins opens new possibilities in drug development and personalized medicine. Rather than relying on traditional trial-and-error methods, researchers can now design proteins specifically tailored to target disease pathways with unprecedented precision. Environmental Solutions The technology's potential extends to environmental challenges, with the ability to design proteins that could efficiently capture carbon dioxide or break down pollutants. This could provide new tools in the fight against climate change and environmental degradation. Materials Science Innovation ESM3's capabilities in protein design extend to the development of new biomaterials, potentially revolutionizing fields from sustainable packaging to advanced electronics. Accelerating Scientific Discovery By compressing evolutionary timescales from millions of years to months, ESM3 dramatically accelerates the pace of biological innovation. This acceleration could lead to breakthroughs in fields that traditionally required extensive trial-and-error experimentation.
The Proof: The GFP Breakthrough
The team demonstrated ESM3's capabilities through a remarkable achievement - creating a novel Green Fluorescent Protein (GFP) that had never existed in nature. While GFPs naturally occur in jellyfish and have been used by scientists as cellular markers, ESM3 designed an entirely new version with unique properties. This achievement effectively demonstrated the model's ability to not just understand but to innovate in protein design.
Looking Ahead: The Future of Biological Engineering
ESM3 represents more than just a technological advancement - it's a fundamental shift in how we approach biological engineering. As researchers and companies begin to explore its capabilities, we're likely to see applications that we haven't even imagined yet. The technology raises important questions about the future of biological engineering and our ability to design life's building blocks. While the potential benefits are enormous, Evolutionary Scale maintains a commitment to responsible development, ensuring that this powerful technology is used ethically and safely. As we stand at this frontier of biological engineering, ESM3 marks a significant step toward making biology programmable - opening up possibilities that were previously confined to the realm of science fiction. Whether it's developing new treatments for diseases, creating sustainable materials, or finding solutions to environmental challenges, ESM3 is poised to play a crucial role in shaping our technological future.
Conclusion
ESM3 represents a convergence of artificial intelligence and biological understanding that could fundamentally transform how we approach some of humanity's biggest challenges. As the technology continues to develop and more researchers and organizations gain access to its capabilities, we're likely to see an acceleration in biological innovation that could reshape multiple industries and scientific fields. Read the full article
#AIFramework#BiologicalProgramming#BiomolecularEngineering#ComputationalBiology#DrugDiscoveryAI#EvolutionaryComputing#GenerativeBiology#MachineLearningBiology#MolecularDesign#ProteinDesignAI#ProteinEngineering#ProteinFolding#ScientificComputing#SyntheticBiology#TherapeuticProteinDesign
0 notes
Text
Genetics PhD student appreciates unnecessary, hostile, and misinformed feedback.
PhD student Janet Small has established a reputation as consistently receptive to, “unnecessary, harsh, and ultimately misguided criticism,” beamed her supervisor. Her colleague, Dan, agrees. “She never argues,” added Dan, nodding. “And she’s always smiling. I like that. I wonder if she’s seeing anybody.” Small, who organizes LGBTAQ+ student gatherings, shared that she expects little from her colleagues and supervisor, whom she perceives as self-interested and unimaginative, but wishes them the best. Adding that she always ignores Dan’s advice, she holds the record for departmental doctoral student publications, a fact she shared while smiling.
#phd l#sciencephd#phdlife#phdproblems#women in science#women in stem#academia#dark academia#feminism#feminist#my awkward academia#stem#women who code#biostatistics#computationalbiology#awkwardacademia#lablife#feedbackloop
4 notes
·
View notes
Text
How MAGNA™ Technology is Revolutionizing the Study of Biomolecular Interactions
In the world of science, understanding how molecules interact with each other is crucial for developing new drugs, diagnostics, and other life-saving technologies. However, traditional methods for studying these interactions are often time-consuming, expensive, and inaccurate.
MAGNA™ technology is a new and innovative approach that is revolutionizing the way we study biomolecular interactions. This powerful tool can measure the strength, structure, and location of interactions between molecules with unprecedented precision and sensitivity.
What is MAGNA™ technology?
MAGNA™ technology is based on a novel method called bioluminescence resonance energy transfer (BRET), It is developed by Depixux. BRET uses light to measure the distance between two molecules. When two molecules that are labeled with special BRET tags come close together, they can transfer energy from one to the other, which produces a light signal. The intensity of the light signal is proportional to the distance between the molecules.
How does MAGNA™ technology work?
MAGNA™ technology can be used in three different modes:
Binding strength mode: In this mode, MAGNA™ is used to measure the force required to pull apart two molecules. This can be used to study the strength of drug-target interactions, protein-protein interactions, and other biomolecular interactions.
Binding structure mode: In this mode, MAGNA™ is used to measure the effect of another molecule on the structure of a target molecule. This can be used to study how drugs bind to their targets, how proteins interact with each other, and how other molecules can alter the structure of biomolecules.
Binding location mode: In this mode, MAGNA™ is used to measure the precise location on a molecule where another molecule binds. This can be used to study the binding sites of drugs, the active sites of enzymes, and other important regions of biomolecules.
What are the benefits of MAGNA™ technology?
MAGNA™ technology offers several advantages over traditional methods for studying biomolecular interactions:
High sensitivity: MAGNA™ can detect interactions between molecules that are too weak to be measured by other methods.
High precision: MAGNA™ can measure the distance between molecules with high accuracy.
Real-time: MAGNA™ can measure interactions in real time, which allows researchers to study the dynamics of biomolecular interactions.
Label-free: MAGNA™ does not require the molecules to be labeled with special tags, which can simplify experiments and reduce costs.
How is MAGNA™ technology being used?
MAGNA™ technology is being used in a variety of research applications, including:
Drug discovery: MAGNA™ is being used to identify new drug targets and to develop new drugs that are more effective and have fewer side effects.
Diagnostics: MAGNA™ is being used to develop new diagnostic tests for diseases such as cancer and Alzheimer's disease.
Protein engineering: MAGNA™ is being used to engineer proteins with new properties and functions.
The future of MAGNA™ technology
MAGNA™ technology is a powerful tool that is revolutionizing the way we study biomolecular interactions. This technology has the potential to lead to the development of new drugs, diagnostics, and other life-saving technologies. As MAGNA™ technology continues to develop by Depixus, we can expect to see even more exciting discoveries in the years to come.
#TechnologyIntroduction#InnovativeTechnology#DepixusTechnology#MAGNATechnology#MAGNAbiomolecularinteractions#MAGNAdrugdiscovery#MAGNAproteinproteininteractions#Biomolecularinteractions#Moleculebinding#Drugdiscovery#Proteinproteininteractions#Biophysics#Structuralbiology#Moleculardynamics#Computationalbiology#depixus
0 notes
Text
How do I get a job in computational biology?
In the career path of this field, you can either take bioscience or any other branch of life science or computer science and engineering at the undergrad level.
One must be fluent in understanding life science terminology of genetics, genomics, and cellular biology.

#genomics#biology#genetics#computerscience#bioscience#computationalbiology#computational#lifescience#student#datascience#programming#jobs#technology#code#machinelearning#hiring#datascientist#developers & startups
0 notes
Text
Computational Biology Market to Grow at 13.6 by 2028: COVID Impact & Post COVID Analysis, Business Opportunities, and Strategies
In 2018, the global computational biology market was valued at USD 2.9 billion, with a CAGR of 21.5 percent predicted over the forecast year. Growing drug discovery R&D, demand for predictive models, use in population-based sequencing efforts like the human genome project, and government financing are all driving the market forward. For example, in March 2019, Ambrx Inc., a clinical-stage biopharmaceutical business specializing in protein therapies, struck a research and development agreement with BeiGene, Ltd., a commercial-stage pharmaceutical company.
According to a new analysis, the global computational biology market is estimated to reach USD 13.6 billion by 2026. During the forecast period, it is expected to grow at a CAGR of 21.3 percent.
read more @ https://bhaokresayali.blogspot.com/2021/08/computational-biology-market-to-grow-at.html
#computationalbiolog marketsize#computationalbiologymarket#biological systems#computationalbiologymarketforecast#Computationalbiology
0 notes