#histone mRNA
Explore tagged Tumblr posts
Text

TSRNOSS, page 209.
#actin#myosin#osmolytes#potassium#ionization potential#prion#denaturing agent#fever#ubiquitin#heat shock proteins#exon shuffling#leucine#zipper proteins#histone mRNA#theoretical biology#manuscript#diaries#satyendra sunkavally
0 notes
Note
Hello! I am a molecular biologist, and I was wondering if I could get your opinion on some of my theories on Gallifreyans.
I haven't read through everything on your blog yet, but I'm working my way through it (lol). So some of this may not be quite accurate with what you have set up thus far.
Basically, I want to briefly discuss alternative splicing! Anyway, in metazoans, alternative splicing outcomes can be regulated in a time and tissue specific manner by legitimately hundreds of biomolecules such as RNA binding proteins, chromatin remodelers, hormones, etc etc. It is subject to epigenetic regulation as alternative splicing and transcription are coupled (and splicing largely occurs cotranscriptionally), so details such as DNA methylation, nucleosome positioning, histone modifications, etc can change the balance of different mRNA isoforms. This is largely because these factors will either help recruit splicing factors (or inhibit their recruitment) or because it will slow RNA Polymerase II elongation.
Onto my theories. I have been thinking for a little while that the lindos hormone can perhaps modulate splicing, triggering the production of regeneration-specific isoforms. Perhaps their bodies work so fast that isoforms promoting totipotency trigger a temporary transition away from the cells' differentiated states.
I also think it could be possible that they have some novel ability to, say, "unsplice," which humans cannot do. This could potentially allow them to use already made transcripts and then completely change them to produce unique proteins without needing to transcribe another mRNA. This could feasibly allow them to rapidly change what proteins are in each cell (perhaps quick enough that it occurs within the regeneration itself). Although, there would be some instability while now unused proteins get degraded or the splicing/unsplicing ratio stabilizes (the molding period). This would require intense regulation as well as unsplicing and resplicing would now be posttranscriptional, but I digress.
Sorry to bother you with the long post, I just had too many nerdy ideas going through my head. Thanks!
-gallifreyanhotfive
Molecular Biology: 'Unsplicing'
Oh, you thrill me with your biology talk! Molecular biology is not a speciality so apologies in advance for any limited response.
🔬 Lindos and Its Variations
Something to be covered in the new Anatomy and Physiology guide is a wider look at the role of Lindos in Time Lords, so we're hitting the nail on the head here.
Under stress, injury, or during the process of regeneration, the lindal gland significantly increases its production of the hormone Lindoneogen like a caffeine-fueled scientist, resulting in a corresponding surge in lindos cell production. There are several forms of lindos cells, including:
Lindopoetic Progenitor Cells (LPCs): Dormant cells that spring into action upon Lindoneogen stimulation.
Lindopoietic Stem Cells (LSCs): Residing in the yellow bone marrow, ready to differentiate under the guidance of Lindoneogen and the catalytic influence of artron, into ...
Lindoblasts and Phagolindotropes: Specialised cells responsible for regenerating tissue and recycling cellular components from the previous incarnation.
Haemolindocytes: Circulating cells that endow Gallifreyan blood with its regenerative properties.
💡Splicing and Lindoneogen
Lindoneogen could play a key role in alternative splicing, creating specific mRNA isoforms vital for regeneration. This implies that Lindoneogen is not just a cellular signal but also a molecular tool for crafting the necessary protein portfolio for regeneration. So Lindoneogen may trigger the production of specific mRNA isoforms that are vital for the regeneration process, which could lead to the expression of proteins that facilitate the transition of cells into a more pluripotent state.
🖇️Unsplicing
Love this idea. 'Unsplicing' as your concept presents would be particularly relevant during regeneration. It could allow cells to quickly alter their protein expression profiles without the lag of new mRNA transcription. This rapid adaptation would be pretty handy for the efficient transition of cells to suit the requirements of the new incarnation.
🔗Integrating with Lindos Cells
This concept of 'unsplicing' could be particularly prominent in the function of phagolindotropes. As these cells are responsible for consuming the previous incarnation’s cells and replacing them with new ones, their ability to 'unsplice' and rapidly change protein expression would be pretty useful. This mechanism might also support the functions of lindoblasts and haemolindocytes in tissue regeneration and blood adaptability.
🏫 So ...
The addition of splicing and unsplicing mechanisms to the lindos theory suggests a more complex and dynamic process than simple cellular proliferation and differentiation, with dynamic genetic adaptations at the molecular level highlighting the advanced biological capabilities of Gallifreyans. Good work, Batman!
More content ... →📫Got a question? | 📚Complete list of Q+A →😆Jokes |🩻Biology |🗨️Language |🕰️Throwbacks |🤓Facts →🫀Gallifreyan Anatomy and Physiology Guide (pending) →⚕️Gallifreyan Emergency Medicine Guides →📝Source list (WIP) →📜Masterpost If you're finding your happy place in this part of the internet, feel free to buy a coffee to help keep our exhausted human conscious. She works full-time in medicine and is so very tired 😴
#doctor who#gil#gallifrey institute for learning#dr who#dw eu#gallifrey#gallifreyan biology#gil biology#gallifreyans#ask answered
21 notes
·
View notes
Text
Exploring the Marvels of Biological Macromolecules: The Molecular Machinery of Life (Part 3)
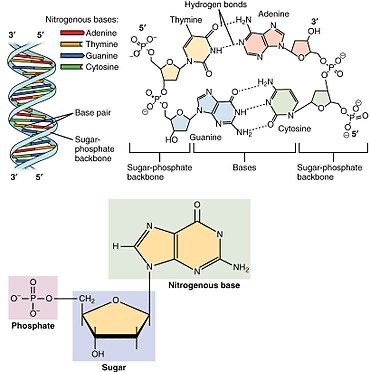
Nucleotide Structure: The Building Blocks
Nucleotides, the monomers of nucleic acids, consist of three fundamental components:
1. Phosphate Group (PO4): Provides a negatively charged backbone for the nucleic acid strand.
2. Pentose Sugar: In DNA, it's deoxyribose; in RNA, it's ribose. The sugar moiety forms the framework of the nucleotide.
3. Nitrogenous Base: Adenine (A), Guanine (G), Cytosine (C), Thymine (T) in DNA, and Uracil (U) in RNA. These bases are responsible for the genetic code.
DNA (Deoxyribonucleic Acid): The Repository of Genes
DNA is a double-stranded helical molecule, with each strand composed of a linear sequence of nucleotides. It encodes the genetic information necessary for an organism's development, growth, and functioning. The Watson-Crick base pairing rules—A with T and C with G
DNA (Deoxyribonucleic Acid): The Repository of Genes
DNA is a double-stranded helical molecule, with each strand composed of a linear sequence of nucleotides. It encodes the genetic information necessary for an organism's development, growth, and functioning. The Watson-Crick base pairing rules—A with T and G with C—ensure DNA's complementary and faithful replication.
RNA (Ribonucleic Acid): From DNA's Blueprint to Protein Synthesis
RNA plays diverse roles in the cell, including serving as a messenger (mRNA) for protein synthesis, a structural component of ribosomes (rRNA), and an adapter molecule (tRNA) that brings amino acids to the ribosome during translation. Unlike DNA, RNA is often single-stranded and contains uracil (U) instead of thymine (T).
Genome Organization and Chromosomes
Genomic DNA is organized into chromosomes within the cell nucleus. These structures enable efficient storage, replication, and transmission of genetic information during cell division and reproduction.
Replication and Transcription
DNA replication ensures the faithful duplication of genetic material during cell division, while transcription converts DNA into RNA, providing a template for protein synthesis.
Translation
The cellular machinery, composed of ribosomes and tRNA, reads the mRNA code and assembles amino acids into polypeptides during translation, ultimately forming functional proteins.
Genetic Code
The genetic code, a triplet code of nucleotide sequences (codons), dictates a protein's sequence of amino acids. It is nearly universal, with only minor variations across species.
Epigenetics
Epigenetic modifications, such as DNA methylation and histone modifications, regulate gene expression without altering the underlying DNA sequence, pivotal in development and cell differentiation.
Macromolecular interactions are the essence of cellular life. Within the complex microcosm of a cell, countless molecules engage in precise and choreographed dances, forming intricate networks that govern every facet of biology. These interactions, governed by the principles of biochemistry, are the foundation upon which life's processes are built.
Amino Acids: The Building Blocks
Proteins are composed of amino acids organic molecules that contain an amino group (-NH2), a carboxyl group (-COOH), a hydrogen atom, and a distinctive side chain (R group). There are 20 different amino acids, each with a unique side chain that confers specific properties to the amino acid.
Primary Structure: Amino Acid Sequence
The primary structure of a protein refers to the linear sequence of amino acids in the polypeptide chain. The genetic information in DNA encodes the precise arrangement of amino acids.
Secondary Structure: Folding Patterns
Proteins don't remain linear; they fold into specific three-dimensional shapes. Secondary structures, such as α-helices and β-sheets, result from hydrogen bonding between nearby amino acids along the polypeptide chain.
Tertiary Structure: Spatial Arrangement
The tertiary structure is the overall three-dimensional shape of a protein, determined by interactions between amino acid side chains. These interactions include hydrogen bonds, disulfide bridges, ionic bonds, and hydrophobic interactions.
Quaternary Structure: Multiple Polypeptide Chains
Some proteins, known as quaternary structures, comprise multiple polypeptide chains. These subunits come together to form a functional protein complex. Hemoglobin, with its four subunits, is an example.
Protein Functions: Diverse and Essential
Proteins are involved in an astounding array of functions:
1. Enzymes: Proteins catalyze chemical reactions, increasing the speed at which reactions occur.
2. Structural Proteins: Proteins like collagen provide structural support to tissues and cells.
3. Transport Proteins: Hemoglobin transports oxygen in red blood cells, and membrane transport proteins move molecules across cell membranes.
4. Hormones: Hormonal proteins, such as insulin, regulate various physiological processes.
5. Immune Function: Antibodies are proteins that play a crucial role in the immune system's defense against pathogens.
6. Signaling: Proteins are critical in cell signaling pathways, transmitting information within cells.
Protein Denaturation and Folding
Protein Diversity: The vast diversity of proteins arises from the combinatorial possibilities of amino acid sequences, secondary structure arrangements, and three-dimensional conformations.
Nucleic acids, the remarkable macromolecules that govern all living organisms' genetic information, are life's quintessential molecules. These complex polymers of nucleotides play an unparalleled role in the storage, replication, and expression of genetic information, shaping the development, characteristics, and functions of every living entity on Earth. Let's embark on an exploration of the intricate world of nucleic acids.
Nucleotide Structure: The Building Blocks
Nucleotides, the monomers of nucleic acids, consist of three fundamental components:
1. Phosphate Group (PO4): Provides a negatively charged backbone for the nucleic acid strand.
2. Pentose Sugar: In DNA, it's deoxyribose; in RNA, it's ribose. The sugar moiety forms the framework of the nucleotide.
3. Nitrogenous Base: Adenine (A), Guanine (G), Cytosine (C), Thymine (T) in DNA, and Uracil (U) in RNA. These bases are responsible for the genetic code.
DNA (Deoxyribonucleic Acid): The Repository of Genes
DNA is a double-stranded helical molecule, with each strand composed of a linear sequence of nucleotides. It encodes the genetic information necessary for an organism's development, growth, and functioning. The Watson-Crick base pairing rules—A with T and G with C—ensure DNA's complementary and faithful replication.
RNA (Ribonucleic Acid): From DNA's Blueprint to Protein Synthesis
RNA plays diverse roles in the cell, including serving as a messenger (mRNA) for protein synthesis, a structural component of ribosomes (rRNA), and an adapter molecule (tRNA) that brings amino acids to the ribosome during translation. Unlike DNA, RNA is often single-stranded and contains uracil (U) instead of thymine (T).
Genome Organization and Chromosomes:
Replication and Transcription: DNA replication ensures the faithful duplication of genetic material during cell division, while transcription converts DNA into RNA, providing a template for protein synthesis.
Translation: The cellular machinery, composed of ribosomes and tRNA, reads the mRNA code and assembles amino acids into polypeptides during translation, ultimately forming functional proteins.
Genetic Code: The genetic code, a triplet code of nucleotide sequences (codons), dictates the sequence of amino acids in a protein. It is nearly universal, with only minor variations across species.
Epigenetics: Epigenetic modifications, such as DNA methylation and histone modifications, regulate gene expression without altering the underlying DNA sequence, pivotal in development and cell differentiation.
Macromolecular interactions are the essence of cellular life. Within the complex microcosm of a cell, countless molecules engage in precise and choreographed dances, forming intricate networks that govern every facet of biology. These interactions, governed by the principles of biochemistry, are the foundation upon which life's processes are built.
#science#biology#college#education#school#student#medicine#doctors#health#healthcare#genetics#genetic engineering#science nerds#dna activation#new dna
24 notes
·
View notes
Text
RNAs, introns and introduction to recombinant DNA.
The genome is a vault of genetic information, but on its own its kind of useless as the information cannot be used in the cell. Utilization of this information requires a coordinated effort from enzymes and proteins, this coordination of chemical reactions is known as genome expression. The initial step of genome expression is known as the transcriptome, a collection of RNA molecules which is made up of the genes active in the cell at the time. (its constantly changing to the environment that the cell is in) the transcriptome is maintained by transcription a simple process in which individual genes are copied and pasted into RNA molecules known as mRNA. The second product within gene expression is known as proteome which you can think of as the cells library of proteins at hand. This gives the cell its unique and individual characteristics that it can express. The proteins in this library known as the proteome are synthesized by the translation of some RNA molecules.
I think it is of utmost importance to start with the most simple and important step…
Transcription:
The first step in transcription is initiation,
Transcription begins with the binding of RNA polymerase 2 to the promoter region of a gene, the most common promotor contains a conserved gene sequence we call the TATA box, but there are many others that exist which are show by the different TF families.
The next step we have the formation of transcription initiation complex, this is when transcription factors such as TATA-binding protein also known as TBP, bind to the promoter, forming a pre-initiation complex. Other general transcription factors join, and RNA polymerase 2 is recruited to form the transcription initiation complex.
Next, we have initiation of transcription where RNA polymerase 2 unwinds the DNA helix at the transcription “start site”. The enzyme catalyses the synthesis of a short RNA primer (which is about 10 nucleotides in length) complementary to the template DNA strand.
The second step is Elongation:
Once the RNA polymerase has synthesized the initial RNA primer, it proceeds to elongate the RNA chain, The RNA polymerase adds ribonucleotide complementary to the template DNA strand. This process is known as RNA chain Elongation.
After this, we have nucleosome remodelling where the DNA in eukaryotic cells is wrapped around histone proteins to form nucleosomes. As transcription proceeds, nucleosomes are temporarily disrupted and then reassembled, allowing RNA polymerase to access the DNA template.
The third step is Termination:
Eukaryotic mRNA molecules undergo post-transcriptional modification, this includes polyadenylation which involves the addition of a poly-A tail to the 3’ end of the RNA. Polyadenylation is then accompanied by cleavage of the RNA precursor at the specific site.
During this process transcription termination signals are recognized by RNA polymerase 2, leading to its dissociation from the DNA template. The termination signal will most likely include the poly-A signal.
The fourth step being RNA processing:
The first step of RNA processing is known as “capping”. Capping the 5’ end of the newly synthesized mRNA is modified with a 7-methylguanosine cap, the whole point of this cap is just to protect the mRNA as it leaves the nucleus and helps in the transportation process.
The second step is splicing, Eukaryotic genes often contain introns (which are kind of gaps or non-coding regions) and exons (which are the important bits, coding regions). In a process called splicing the annoying useless introns are cut and the exons are then pinched together with each other. This is all catalysed by a complex of RNA and proteins known as spliceosomes.
The fifth step being transport and translation:
This is when mRNA is transported out of the nucleus to the cytoplasm where it serves as a template for translation.
The sixth step is regulation:
Gene expression if tightly regulated at the transcriptional level by various elements, including transcriptional factors and chromatin modification, there is also enhancers and silencers that help regulate elements that influence transcription.
We also have post-transcriptional regulation, which is processes such as alternative splicing and RNA stability which play roles in determining the final mRNA products.
It is important to acknowledge that Eukaryotic transcription is more complex than prokaryotic transcription due to the presence of introns, the involvement of multiple RNA polymerases, and the necessity for additional processing steps.
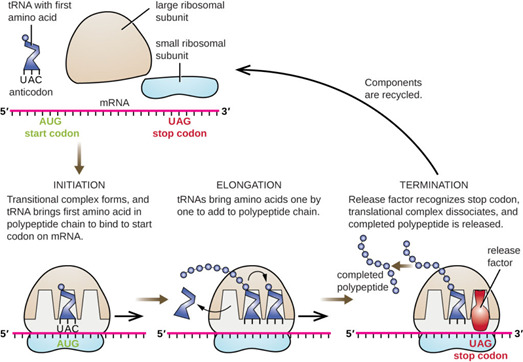
This diagram helps visualise the main steps more simply.
Hope this was clear excuse any mistakes in grammar :)
Referencing:
Picture link above^
2 notes
·
View notes
Text
Substansi Genetik : DNA, Kromosom, dan Sintesis Protein
Pendahuluan Materi genetik merupakan unsur penting yang menentukan sifat biologis semua makhluk hidup.Informasi genetik ini disusun dalam bentuk DNA dan disimpan dalam struktur yang disebut kromosom. Proses sintesis protein mengubah informasi genetik menjadi fungsi biologis yang penting. Artikel ini memberikan pandangan mendalam tentang sintesis DNA, kromosom, dan protein, serta menjelaskan pentingnya memahami materi genetik dalam ilmu kehidupan modern.
DNA: Informasi Genetik Molekul Asam deoksiribonukleat (DNA) adalah molekul yang membawa informasi genetik yang diperlukan untuk perkembangan, fungsi, pertumbuhan, dan reproduksi semua organisme hidup dan banyak virus. DNA memiliki dua heliks yang melilit satu sama lain untuk membentuk struktur heliks ganda. Setiap heliks terdiri dari nukleotida yang terdiri dari tiga komponen utama: gula deoksiribosa, fosfat, dan basa nitrogen. Ada empat jenis basa nitrogen dalam DNA: adenin (A), timin (T), sitosin (C), dan guanin (G). Urutan basa ini menentukan kode genetik yang menyimpan informasi tentang pembentukan protein.
Kromosom: Struktur Penyimpanan DNA DNA pada sel eukariotik tersusun dalam kromosom di dalam inti sel. Manusia mempunyai 23 pasang kromosom. Artinya, setiap sel dalam tubuh manusia memiliki total 46 kromosom. Kromosom tersusun dari DNA yang dibungkus protein yang disebut histon, membentuk struktur padat dan terorganisir. Struktur ini penting untuk menjaga stabilitas gen dan mengendalikan ekspresi gen.
Sintesis Protein: Transkripsi dan Translasi Proses sintesis protein melibatkan dua langkah utama: transkripsi dan translasi.
Transkripsi: Proses ini dimulai dengan pengikatan enzim RNA polimerase pada DNA di wilayah yang disebut promotor. RNA polimerase membuka heliks ganda DNA dan menggunakan satu untai sebagai cetakan untuk sintesis messenger RNA (mRNA). mRNA adalah salinan kode genetik yang diperlukan untuk membuat protein. Setelah transkripsi selesai, mRNA meninggalkan inti sel dan berpindah ke ribosom di sitoplasma.
Translasi: Di ribosom, mRNA dibaca oleh ribosom dalam kelompok tiga basa yang disebut kodon. Setiap kodon spesifik untuk asam amino tertentu. Molekul RNA transfer (tRNA) mengangkut asam amino yang sesuai ke ribosom, di mana asam amino tersebut dirangkai menjadi rantai polipeptida sesuai dengan urutan kodon pada mRNA. Rantai polipeptida ini terlipat menjadi struktur tiga dimensi yang membentuk protein fungsional.
Fungsi protein di dalam sel Protein adalah molekul sangat penting yang memiliki berbagai fungsi di dalam sel. Beberapa protein berfungsi sebagai enzim yang mempercepat reaksi biokimia, sementara yang lain berfungsi sebagai protein struktural yang mendukung dan membentuk sel. Beberapa protein berperan dalam mengatur aktivitas gen dan sinyal sel. Misalnya saja hemoglobin merupakan protein dalam sel darah merah yang membawa oksigen ke seluruh tubuh, dan hormon insulin merupakan protein yang mengatur kadar gula darah.
Regulasi Ekspresi Gen Regulasi ekspresi gen adalah proses mengendalikan kapan dan di mana gen untuk sintesis protein diaktifkan. Peraturan ini penting untuk memastikan bahwa protein yang tepat diproduksi pada waktu dan tempat yang tepat, tergantung kebutuhan sel atau organisme. Regulasi ekspresi gen terjadi pada berbagai tingkatan, termasuk transkripsi, pemrosesan mRNA, translasi, dan pasca-translasi.
Penelitian dan Penerapan Studi tentang materi genetik telah menghasilkan banyak penemuan penting dalam biologi dan kedokteran. Misalnya, teknologi CRISPR-Cas9 memungkinkan para ilmuwan mengedit gen secara spesifik, sehingga membuka kemungkinan untuk mengobati penyakit genetik. Selain itu, penelitian tentang sintesis DNA dan protein telah membantu kita memahami mekanisme dasar kehidupan dan evolusi serta membuka jalan bagi pengembangan obat-obatan baru dan terapi gen.
Kesimpulan Pemahaman materi genetik, seperti DNA, kromosom, dan sintesis protein, merupakan dasar biologi modern. Informasi ini tidak hanya penting untuk memahami perkembangan dan fungsi organisme hidup, namun juga mempunyai implikasi besar bagi bioteknologi dan kedokteran. Seiring dengan berlanjutnya penelitian, kita dapat belajar lebih banyak tentang cara kerja gen dan bagaimana pengetahuan tersebut dapat digunakan untuk meningkatkan kesehatan dan kualitas hidup manusia.
1 note
·
View note
Text
Persistent Spike Protein Production and Early Mortality: Exploring the Detrimental Impact of Frameshift Mutations in mRNA Vaccines

In the rapidly evolving landscape of mRNA vaccines, understanding the mechanisms underlying persistent spike protein production and potential adverse effects is paramount. Emerging scientific theories suggest that persistent spike protein production, frameshift mutations, and related mechanisms could lead to earlier mortality in some individuals. Here's a comprehensive exploration of these phenomena:
Persistent Spike Protein Production Mechanisms
Integration into the Genome Emerging evidence suggests that mRNA from vaccines could potentially undergo reverse transcription and integrate into the host genome. Although traditional understanding posits that mRNA remains in the cytoplasm, some studies indicate that under specific conditions, integration might occur. This could happen through:
Reverse Transcriptase Presence: Reverse transcriptase enzymes from other infections or cellular sources may facilitate the reverse transcription of vaccine mRNA, resulting in integration into the host DNA. Nuclear Entry: Under certain conditions, such as inflammation or cellular stress, mRNA might gain access to the nucleus.
Epigenetic Modifications mRNA vaccines could potentially induce long-lasting epigenetic changes that sustain spike protein production. This could be due to:
Immune Response-Induced Changes: Prolonged alterations in gene expression patterns could result from vaccine-induced immune responses. Cellular Stress: The stress induced by the vaccine formulation or immune response might lead to epigenetic modifications that continue to drive spike protein production. Histone Modifications and DNA Methylation: Changes in histone acetylation or DNA methylation could result in sustained activation of spike protein-encoding genes.
Viral Reservoirs A proposed mechanism involves the establishment of viral reservoirs in specific tissues, where spike protein production could persist:
Localized Immune Responses: The vaccine may provoke localized immune reactions that lead to sustained spike protein expression in certain tissues. Immune Privilege Sites: Some tissues, such as the central nervous system or reproductive organs, may serve as immune-privileged sites where spike protein production persists due to limited immune surveillance.
Circulating Spike Protein Research has revealed elevated levels of circulating spike protein in individuals experiencing adverse events post-vaccination, such as myocarditis. This phenomenon could be due to:
Inflammatory Responses: Inflammation might prolong the presence of spike protein in the bloodstream. Autoimmune Phenomena: Autoimmune reactions could also contribute to persistent spike protein production.
Potential Dysregulation There are indications that certain immune responses or conditions might lead to continued spike protein production even after mRNA degradation. This dysregulation could result from:
Lipid Nanoparticles: The lipid nanoparticles used in vaccine formulations might trigger inflammatory responses that interfere with the degradation of mRNA. Spike Protein-Host Cell Interactions: Interactions between the spike protein and host cellular machinery could lead to prolonged spike protein production.
Understanding Frameshift Mutations and Their Negative Effects Recent research has highlighted that modifications like 1-methyl-Ψ (1-methylpseudouridine) enhance mRNA stability and efficacy but may also increase the production of frameshifted proteins. Frameshift mutations are known for their detrimental effects, often leading to severe clinical outcomes:
Loss of Function: Frameshift mutations result in premature stop codons, leading to truncated and non-functional proteins. Such mutations can impair essential biological functions, particularly in critical proteins like enzymes or structural proteins. Gain of Toxic Function: In some cases, frameshift mutations produce elongated or misfolded proteins that gain aberrant, toxic functions, contributing to cellular dysfunction. Disease Association: Frameshift mutations are implicated in various genetic disorders and cancers. For instance, they can cause conditions like cystic fibrosis or muscular dystrophy, where normal protein function is disrupted. Cellular Stress and Apoptosis: The production of abnormal proteins can trigger the unfolded protein response (UPR) and endoplasmic reticulum (ER) stress, potentially leading to apoptosis (programmed cell death) or contributing to neurodegenerative diseases. Immune Response to Aberrant Proteins: Mice vaccinated with the BNT162b2 mRNA vaccine (Pfizer-BioNTech) exhibited heightened immune responses against frameshifted products compared to those vaccinated with viral vector vaccines. This immune response to aberrant proteins could have implications for both efficacy and adverse reactions.
Early Mortality Concerns Scientific theories suggest that persistent production of spike proteins, coupled with frameshift mutations, may contribute to increased early mortality among individuals who received doses of mRNA vaccines. This risk could vary depending on dosage and individual body response, particularly due to:
Frameshift Mutations Leading to Dysfunctional Proteins: The production of dysfunctional proteins due to frameshift mutations could result in chronic cellular stress and disease progression.
Persistent Spike Protein Production and Early Mortality Concerns
Persistent Spike Protein Production: Prolonged spike protein production, whether due to genomic integration, viral reservoirs, or epigenetic modifications, could lead to chronic inflammation or autoimmune reactions. This persistent antigenic presence may: Trigger Chronic Inflammation: Continuous immune activation can lead to tissue damage, fibrosis, and organ dysfunction. Induce Autoimmune Reactions: Persistent spike protein expression may break immune tolerance, leading to the development of autoimmune diseases. Frameshift Mutations and Immune Dysregulation: Aberrant Immune Responses: Frameshift mutations could produce neoantigens that the immune system recognizes as foreign, potentially leading to immune-mediated tissue damage. Cytokine Storms: The immune response to persistent spike proteins may result in hyperinflammatory states such as cytokine storms, further contributing to organ damage and earlier mortality. Amplifying Adverse Effects through Frameshift Mutations Increased Production of Aberrant Proteins: 1-Methyl-Ψ Modifications: While enhancing mRNA stability and efficacy, these modifications may also increase the likelihood of frameshift mutations, leading to a higher production of aberrant proteins. Ribosomal Slippage: Errors in reading frames due to ribosomal slippage can exacerbate the production of dysfunctional proteins. Impaired Protein Quality Control: Proteasomal Overload: A surge in aberrant proteins may overwhelm the proteasome, impairing its ability to degrade misfolded proteins. ER Stress and UPR Activation: Accumulation of misfolded proteins in the endoplasmic reticulum can trigger the unfolded protein response, leading to ER stress and apoptosis. Disease Progression and Early Death: Neurodegenerative Diseases: Persistent cellular stress and aberrant protein accumulation are known contributors to neurodegenerative diseases like Alzheimer's and Parkinson's. Cardiac Complications: Chronic inflammation and immune dysregulation can lead to myocarditis, pericarditis, and other cardiac conditions. Cancer Development: Frameshift mutations and immune dysregulation could increase the risk of oncogenesis by promoting genomic instability.
Conclusion While mRNA vaccines represent a remarkable scientific breakthrough, it is crucial to investigate the persistent spike protein production mechanisms and potential frameshift mutations that might contribute to earlier mortality among some individuals. Further research into these mechanisms will be essential for understanding the long-term safety profile of mRNA vaccines and ensuring their safe and effective use in the future.
Long-Term Manifestation of Harmful Effects Given the nature of frameshift mutations and their impact on essential biological functions, harmful effects could manifest or continue to manifest long after initial exposure to mRNA vaccines. Factors that could contribute to long-term adverse outcomes include:
Accumulation of Truncated or Abnormal Proteins: Continuous production of dysfunctional proteins due to frameshift mutations may lead to cumulative cellular damage over time. Persistent Spike Protein Production: Prolonged spike protein production, whether due to genomic integration, viral reservoirs, or epigenetic modifications, could lead to chronic inflammation or autoimmune reactions. This persistent antigenic presence may: Trigger Chronic Inflammation: Continuous immune activation can lead to tissue damage, fibrosis, and organ dysfunction. Induce Autoimmune Reactions: Persistent spike protein expression may break immune tolerance, leading to autoimmune diseases that could manifest years later. Frameshift Mutations Leading to Dysfunctional Proteins: Critical Proteins Affected: Frameshift mutations could impair essential biological functions in critical proteins like enzymes, structural proteins, and those involved in DNA repair and cell cycle regulation. Chronic Cellular Stress: The accumulation of abnormal proteins may cause prolonged ER stress, unfolded protein response (UPR) activation, and programmed cell death (apoptosis). This cellular stress could contribute to neurodegenerative diseases and other chronic conditions. Increased Risk of Neurodegenerative Diseases: Protein Misfolding and Aggregation: Frameshift mutations and persistent spike protein production could lead to the misfolding and aggregation of proteins, which is a hallmark of neurodegenerative diseases like Alzheimer's, Parkinson's, and Huntington's. Neuroinflammation: Sustained immune activation within the central nervous system could exacerbate neuroinflammation, accelerating neurodegeneration. Cardiac Complications: Myocarditis and Pericarditis: Persistent spike protein production may lead to chronic inflammation of the heart muscle (myocarditis) and outer lining (pericarditis), potentially resulting in long-term cardiac complications. Accelerated Atherosclerosis: Chronic inflammation could contribute to the development and progression of atherosclerosis, increasing the risk of cardiovascular events. Oncogenesis and Cancer Development: Genomic Instability: Frameshift mutations in genes involved in DNA repair and cell cycle regulation could lead to genomic instability and an increased risk of cancer. Chronic Inflammation and Cancer: Persistent spike protein production may result in chronic inflammation, which is a known promoter of tumorigenesis. Immune Dysregulation and Autoimmune Diseases: Autoimmune Phenomena: Frameshift mutations in genes regulating immune tolerance could increase susceptibility to autoimmune diseases. Cytokine Storms: Aberrant immune responses due to persistent spike protein production could lead to hyperinflammatory states like cytokine storms, which could have long-term health implications.
0 notes
Text
The Role of Epigenetic Modifications in Gene Regulation: A Critical Review Cellular differentiation, development, and response to various environmental changes all are multistep biological processes that depend fundamentally on gene regulation. Epigenetic modifications allow the cell to respond interactively to environmental changes without altering the DNA sequence and have thus gained increased attention during the last few years. This blog will critically review some of the important epigenetic mechanisms-DNA methylation, histone modifications, and noncoding RNAs-and their involvement in health and disease.
What is Epigenetic? Epigenetics is defined as the study of heritable features concerning the role of gene regulation and not the underlying DNA sequence. These types of modification, usually reversible, have the ability to dynamically alter the expression of a gene by a cell. The epigenetic mechanisms allow cell differentiation in multicellular organisms beginning from the same DNA sequence genome.
Key Mechanisms of Epigenetic Regulation
DNA Methylation
DNA Methylation DNA methylation is one among the well-studied epigenetic mechanisms. It comprises the addition of a methyl group to the cytosine residue in the CpG dinucleotides, usually associated with the silencing of gene expression. Methylation patterns are important in normal development, while abnormal methylation is associated with diseases including cancer. For instance, the hypermethylation of tumor suppressor genes has been related to the inactivation of such genes in many kinds of cancers Esteller 2007.
Histone Modifications
DNA wraps around core histone proteins to form a nucleoprotein called chromatin, and changing the tail domains of the histones affects both the structure of chromatin and gene expression. Acetylation, methylation, phosphorylation, and sumoylation of histones affects the access of DNA to the transcriptional machinery. For example, in general, histone acetylation activates transcription whereas deacetylation causes silencing, Kouzarides 2007.
Non-coding RNAs: ncRNAs Non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), also play significant roles in gene regulation. miRNAs can degrade messenger RNA (mRNA) or inhibit translation, thus controlling gene expression post-transcriptionally. lncRNAs, on the other hand, regulate gene expression at various levels, including chromatin modification, transcription, and post-transcriptional processes (Rinn & Chang, 2012).
Critical Review: Epigenetic Changes Are of a Janus Nature Epigenetic modification is an indispensable component in physiological processes but turns out to be a double-edged sword since inappropriate epigenetic change gives rise to diseases like tumorigenesis, neurodegeneration, and autoimmune disease.
Epigenetic Therapies Epigenetic alterations, due to their reversible nature, represent very promising therapeutic targets. Therapeutic drugs like DNA methyltransferase inhibitors (for example, azacitidine) and histone deacetylase inhibitors (for example, vorinostat) are effective in the treatment of some cancers (Jones et al., 2016). However, the main challenges lie in guaranteeing specificity because broad epigenetic reprogramming leads to off-target effects.
Epigenetic and Environmental Influence The epigenome is very sensitive to nutrition, toxicants, and stresses. For example, prenatal exposure to malnutrition results in epigenetic modification, increasing the susceptibility to metabolic diseases later in life (Heijmans et al., 2008). Such sensitivities underline how understanding of epigenetic regulation involves lifestyle factors.
Conclusion:
Epigenetic modifications also play crucial roles in fine-tuning gene expression and the maintenance of cellular homeostasis. While allowing new possibilities of therapeutic intervention, especially in cancer, some problems remain to be investigated concerning specificity and also environmental impact. Further research will thus become necessary regarding the dynamic nature of the epigenome if specific and efficacious treatments are to be developed.
References
Esteller, M. (2007). Epigenetic gene silencing in cancer: The DNA hypermethylome. Human Molecular Genetics, 16(R1), R50–R59.
Heijmans, B. T., Tobi, E. W., Stein, A. D., Putter, H., Blauw, G. J., Susser, E. S., Slagboom, P. E., & Lumey, L. H. (2008). Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences, 105(44), 17046-17049.
Jones, P. A., Issa, J. P., & Baylin, S. (2016). Targeting the cancer epigenome for therapy. Nature Reviews Genetics, 17(10), 630–641.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell, 128(4), 693-705.
Rinn, J. L., & Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annual Review of Biochemistry, 81, 145-166.
1 note
·
View note
Text
Ayurveda And Epigenetics
Ayurveda is the oldest, most comprehensive, and natural health care system that originated in India in Vedic times. It is the “science of life” and was used as the main language in communication and teaching in ancient Vedic times. It studies the human body in depth which leads to knowledge about human behavior, physical characteristics, emotional balances, genetic material, composition, and the person as a whole.
DNA is the entity whose presence distinguishes between living and nonliving things. Further, the differences between the living things are observed due to differences in the order of the bases which are, adenine, thymine, guanine, and cytosine, present in DNA. The combination of DNA makes up genes. The human body is estimated to have 2000 genes and only 2% of these genes express themselves and are known as genotypes. The part of genetic material that determines a specific characteristic of an individual is known as a genotype. The part of genetic material that refers to the physical property of an individual, which includes appearance, development, and behavior is known as phenotype.
Epigenetics refers to the external modification of DNA that turns genes on and off, affecting gene expression, that is, by changing the phenotype of the genes and not genotype. This modification takes place without a change in the DNA sequence. This modification in genetic expression leads to transgenerational effects. The major factors that may cause epigenetic changes are lifestyle, behavior, diet, stress, digestion, and other environmental factors. Thus these factors affect the Prakriti of an individual, which refers to phenotype, and indirectly the Janma (birth) Prakriti, which corresponds to genotype. Thus epigenetics become an important mechanism of Ayurveda in understanding the Prakriti of an individual.
DNA methylation, histone modification, chromatin remodeling, and microRNA are the mechanisms that are involved in the modification of DNA expression.
1.In the DNA methylation process, methyl groups are attached to DNA molecules. Due to this the activity e of the DNA changes.
2.The proteins that are wrapped around DNA in the nucleus forming chromatin, are known as histones. In this process, DNA is condensed into a more compact form to protect DNA structure and sequence. This condensation is possible because chromatin can condense or relax, thereby changing the expression of DNA. This condensation and relaxation process of chromatin is supported by histone and affects DNA expression.
3.The rearrangement of chromatin from condensed state to a transcriptionally accessible state, is known as chromatin remodeling. This allows transcription factors to access DNA and control gene expression.
4.The non-coding RNA molecules that stop the functioning of mRNA are referred to as microRNA.
Over 90% of life is controlled by epigenetics which are changes in gene expression. These are brought by what one does in one’s life. Whatever an individual does in its life, it changes the ayurvedic deha prakriti (psychological constitution), which is relayed back to DNA, and changes the expression accordingly. Thus, the process of epigenetics refers to Karma (action) at the cellular level. It can also be referred to Newton’s third law- ” every action has an equal and opposite reaction”.
The four main factors that affect an individual’s life are the same as those mentioned in Ayurveda for maintenance of healthy life and prevention of disease.
1.Lifestyle and behavior
2.Diet and digestion
3.Stress
4.Environmental factors.
If all the actions performed by an individual are in a positive direction, Deha Prakriti is balanced and health is maintained. But if the actions performed by an individual are in a negative direction, Deha Prakriti becomes imbalanced, leading to Vikriti, and disease is manifested. This entire process is brought in by the mechanism of epigenetics.
The healthcare system in Ayurveda lays down a comprehensive text that relates to all the above factors which affect epigenetics.
1.Dinacharya: It recommends the optimal time for waking up, eating, exercising, meditating, and having lunch; in order to stay in sync with the rhythm of the external environment, and supports optimal health.
2.Ratricharya: It requires what to eat for dinner at the optimal time to go to bed in order to get good sleep.
3.Ritucharya: Its recommendations include what all stuff can be consumed in which season. For example, having cool foods during summer to maintain body temperature and minimize hot and spicy food.
Other health-promoting systems in Ayurveda are:
1.Aahara Vihara – diet and guidelines for eating.
2.Sadvritta – Social and personal behavior.
3.Manasa Tivra – Mental stress management.
4.Manas Vritti – Mental fluctuations management.
5.Paryavarana – External and home place management.
On a concluding note, the term Janma Prakriti refers to genotype and Deha Prakriti refers to phenotype. An imbalance or disorder in Deha Prakriti is known as Vikriti and corresponds to disease in the medical system. There are various factors that affect the Prakriti of an individual, and Ayurveda has a cure to it. Thus, the deep knowledge of Ayurveda promotes healthy life even at genetic level.

0 notes
Text
Toxics, Vol. 12, Pages 296: Decreased Ubiquitination and Acetylation of Histones 3 and 4 Are Associated with Obesity-Induced Disorders of Spermatogenesis in Mice
Background: Obesity, a chronic metabolic disorder, is related to cardiovascular diseases, diabetes, #cancer, and reproductive disorders. The relationship between obesity and male infertility is now well recognized, but the mechanisms involved are unclear. We aimed to observe the effect of obesity on spermatogenesis and to investigate the role of histone ubiquitination and acetylation modifications in obesity-induced spermatogenesis disorders. Methods: Thirty male C57BL/6J mice were randomly divided into two groups. The control group was fed with a general maintenance diet (12% fat), while a high-fat diet (HFD) group was fed with 40% fat for 10 weeks; then, they were mated with normal females. The fertility of male mice was calculated, testicular and sperm morphology were observed, and the expression levels of key genes and the levels of histone acetylation and ubiquitination modification during spermatogenesis were detected. Results: The number of sperm was decreased, as well as the sperm motility, while the number of sperm with malformations was increased. In the testes, the #mRNA and protein expression levels of gonadotropin-regulated testicular #RNA helicase (GRTH/DDX25), chromosome region maintenance-1 protein (CRM1), high-mobility group B2 (HMGB2), phosphoglycerate kinase 2 (PGK2), and testicular angiotensin-converting enzyme (tACE) were decreased. Furthermore, obesity led to a decrease in ubiquitinated H2A (ubH2A) and reduced levels of histone H3 acetylation K18 (H3AcK18) and histone H4 acetylation K5, K8, K12, and K16 (H4tetraAck), which disrupted protamine 1 (Prm1) deposition in testis tissue. Conclusion: These results suggest that low levels of histone ubiquitination and acetylation are linked with obesity-induced disorders during spermatogenesis, contributing to a better understanding of obesity-induced damage to male reproduction. https://www.mdpi.com/2305-6304/12/4/296?utm_source=dlvr.it&utm_medium=tumblr
0 notes
Text
Epigenetics and Autoimmunity: How Lifestyle Choices Impact Gene Expression
In the realm of functional medicine, understanding the intricate interplay between genetics and lifestyle is crucial. Epigenetics, the study of changes in gene expression without alterations in the DNA sequence, has gained significant attention for its role in autoimmune conditions.
Understanding how lifestyle factors influence the activity of protein coding genes enables us as practitioners to craft personalized interventions to harness the body's innate potential for healing and gene regulation.
The Epigenetic Landscape
Epigenetics provides a new perspective on the old nature vs. nurture debate. It revolves around changes in gene expression influenced by internal and external factors, such as diet, stress, cell cycle, toxins, immune cells, and physical activity. These changes in gene transcription are carried out by mechanisms like DNA methylation, histone modifications, and the action of regulatory proteins.
In autoimmune diseases, epigenetic modifications can trigger the immune system to attack the body's own tissues. This intricate dance of eukaryotic gene expression showcases the dynamic interplay between genes and their environment
Decoding Gene Expression
Gene expression orchestrates the functioning of our bodies by dictating when and how our genes produce regulatory proteins and other molecules. It's a dynamic process influenced not only by our genetic makeup but also by external factors like lifestyle choices.
This intricate mechanism involves the transcription of genetic information into messenger RNA (mRNA) and subsequent translation into proteins, which are the building blocks and functional molecules essential for cellular processes. Yet, the remarkable aspect of genetic activity lies in its adaptability—a feature that allows our bodies to respond to changing internal and external cues.
https://drritamarie.com/blog/how-lifestyle-choices-impact-gene-expression/
#GeneExpression#LifestyleChoices#HealthGenetics#Epigenetics#WellnessJourney#HealthyHabits#HolisticHealth#MindBodyConnection#OptimalLiving#WellnessWisdom#GeneticWellbeing#HealthyLifestyle#WellnessChoices#EpigeneticFactors#WellnessScience
0 notes
Text
Unlocking the Mysteries of Transcriptional Regulation
Transcriptional regulation lies at the core of gene expression, enabling cells to respond aptly to both internal and external cues. Gaining insights into the molecular intricacies of this process is crucial for unraveling the complexities of life and its myriad processes. This article delves deep into the realm of transcriptional regulation, shedding light on its mechanisms and significance.
Transcription, the initial step of gene expression, orchestrates the conversion of genetic information from DNA to RNA. At the heart of this process lies RNA polymerase, the enzyme responsible for reading the DNA sequence and synthesizing a complementary RNA strand.
Promoters, specific DNA sequences, act as beacons for RNA polymerase during transcription initiation. Additionally, enhancers play a role in fine-tuning transcription rates by interacting with distal regulatory elements.
Central to the control of transcription are transcription factors – proteins that bind to specific DNA sequences. These factors are categorized into activators and repressors, each exerting distinct effects on gene expression.
Forming a complex regulatory network, transcription factors interact with promoters and enhancers, ultimately determining the level of gene expression. Their actions can be influenced by various cellular signals and environmental cues.
A dynamic interplay between activators and repressors allows genes to be activated or repressed in response to cellular needs, fine-tuning gene expression.
Feedback loops are integral to transcriptional regulation, ensuring the maintenance of cellular homeostasis. Negative feedback counteracts changes, while positive feedback reinforces cellular responses.
Epigenetic changes, like histone modifications, significantly impact transcriptional regulation. Chromatin structure alterations brought about by acetylation, methylation, phosphorylation, and more, dictate gene accessibility.
A critical epigenetic modification, DNA methylation, can silence gene expression. These methylation patterns are heritable and can be influenced by various factors, including environmental stimuli.
Following transcription, RNA undergoes splicing, where introns are removed, and exons are joined to create mature mRNA. Alternative splicing allows for multiple protein isoforms to arise from a single gene.
The stability of mRNA molecules plays a vital role in their availability for translation. Several factors can influence mRNA half-life, consequently affecting overall gene expression levels.
Cells possess the remarkable ability to alter gene expression patterns in response to external stimuli such as stress, temperature changes, or exposure to specific chemicals. This adaptive response enables cells to survive and function under diverse conditions.
Stress and nutrition significantly impact transcriptional regulation. Hormones released during stress and nutrients in the diet influence gene expression, thereby affecting overall cellular function.
During development, cells undergo differentiation, acquiring specific functions. Transcriptional regulation plays a pivotal role in determining cell fate and specialization.
Unique sets of genes are expressed in different tissues and organs, orchestrated by tissue-specific transcription factors. This diversity underpins the formation of specialized structures and functions in multicellular organisms.
In the context of cancer, dysregulated gene expression is a common occurrence. Oncogenes and tumor suppressor genes are frequently affected, leading to uncontrolled cell growth and tumor formation.
Disruptions in transcriptional regulation can contribute to neurological disorders, impacting brain development and function. Understanding these mechanisms may provide insights into potential therapeutic strategies.
Therapeutic interventions can be devised to target transcriptional regulators, thereby modulating gene expression to treat various diseases.
Gene therapy, including the revolutionary CRISPR-Cas9 technology, holds the promise of correcting genetic disorders through modification of transcriptional regulation.
Unfaltering research in genomics and epigenetics continues to unveil novel facets of transcriptional regulation, paving the way for groundbreaking discoveries.
The comprehension of transcriptional regulation holds the potential to revolutionize medicine and our understanding of life itself. From personalized medicine to regenerative therapies and beyond, this sophisticated and dynamic process stands at the forefront of scientific progress.
0 notes
Text
This isn't my field (genetics is a major part of Bioengineering, but only a part - not the whole) but our environment (pollutants, diet, physical and mental stress, etc.) is able to alter gene expression. It does so reversibly via DNA methylation (if a methyl group is added it prevents transcription of the sequence it's bound to into mRNA) or histone (the protein that DNA wraps around) modification (causing the DNA to wrap either tighter (inhibits expression) or looser (DNA can be more easily read) around the histone). Since it doesn't change the DNA sequence itself, the effects are reveisible but the epigenetic factors causing the effect seem to be heritable as you said, though it's still being studied to understand exactly why / how.
This is a really comprehensive review if you want to read up on it: Yehuda, R. and Lehrner, A. (2018). Intergenerational transmission of trauma effects: Putative role of epigenetic mechanisms. World Psychiatry, [online] 17(3), pp.243–257. doi:https://doi.org/10.1002/wps.20568.
A famous example that kickstarted studies of famine-related epigenetics was the Dutch Famine / Hunger Winter from 1944-1945 (Heijmans, B.T., Tobi, E.W., Stein, A.D., Putter, H., Blauw, G.J., Susser, E.S., Slagboom, P.E. and Lumey, L.H. (2008). Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences, 105(44), pp.17046–17049. doi:https://doi.org/10.1073/pnas.0806560105.
Even after 6 generations IGF2 expression remained lower than people who's ancestors didn't starve in the famine / who's grandparents weren't prenatally exposed to the effects of famine. It also seems to indicate that epigenetic changes in early development have a greater impact that those later in life (given the persistance of the changes here, though I'm not sure if that's confirmed by other studies).
If my theory of Fowler being in his late 60s or early 70s in BES is true, then he would've been but a wee lad during the famine, so no wonder he's a bit of a "dumpling". I can't say if any epigenetic changes play a part in his peculiarities, partly because I don't personally subscribe to the notion that kinks are born only from trauma, but they can be.
Sorry to blab, I just like talking about stuff like this! ^^
"Only a master could make that cut"

I've seen a few people on Reddit and Youtube talk about how Fowler was supposedly hyped up as a master swordsman and were thus either disappointed by Mizu and Fowler's fight in episode 8 OR they felt it didn't fit Fowler and that he was made OP. Starkly different assessments.
Personally, every scene of or discussing Fowler (namely how Madame Kaji speaks of him) points towards his main assets (combat wise) being strength and brutality.
I personally think Mizu's assumption that only a Master could make that cut makes sense for how Mizu learnt swordplay - namely by practicing what she observed masters of various schools perform for Master Eiji as part of their sword being forged appropriately for how they'd use it. Taigen somewhat challenges Mizu's idea that skill matters most in combat (since her skill and wit is proven effective more often than not) by overpowering Mizu despite using Shindo Ryu (trash technique). Showing that strength can be as important as skill in turning the tide.
The Giant reinforces the power of strength and Mizu's difficulty outperforming it in combat. This is then reinforced by Taigen's comment about The Giant not needing a weapon to kill her (which The Giant almost does in episode 6).
Plus, the Tea Party fight showcases ways Fowler can (and then does) overpower Mizu. Including a chokehold, and even breaking her bones in the same hold as Fowler.


Mizu is literally and figuratively bringing a knife to a gun fight. Fowler doesn't need to have mastered the sword, as Mizu has, to beat Mizu.


Basically, I don't think Mizu's comment about the flower is enough to assume Fowler is a master swordsman. Given that we see him before Mizu does, we see what he is actually like and capable of. That cut was made cleanly because Fowler is both strong and precise. A practiced sadist with two decades of experimentation to hone any method of injury he pleases, not a swordsman.
267 notes
·
View notes
Text
Going through questions:
The genetic code is degenerate because more than one codon can code for 1 amino acid. Every tRNA goes with a specific amino acid; due to the degeneracy of the genetic code, the tRNA will respond to whichever codons go with its specific amino acid. The wobble hypothesis is that the third nucleotide doesn't have to do traditional base pairing.
Bloom syndrome is an autosomal recessive mutation in the BLM gene. It causes defect of helicase and presents with growth retardation, photosensitivity, immunodeficiency, and microcephaly.
Lyonization = X inactivation; in females, one of the X chromosomes is methylated and becomes a Barr body. This inactivated X chromosome becomes tightly would heterochromatin that isn't expressed. Heterochromatin = methylated DNA +deacetylated histones. I recall from listening to OnlineMedEd that acetylation makes DNA accessible. So deacetylation makes DNA less accessible. DNA is wrapped around histones. Methylation of DNA and deacetylation of histones makes the DNA less accessible for transcription. Euchromatin is not methylated and is easily accessible.
Precursor mRNA (pre-mRNA, aka heterogeneous nuclear RNA, hnRNA) is processed before leaving the nucleus--it gets the 5' cap and poly A tail added and introns are spliced out. Once all that happens, it's mature mRNA, which can leave the nucleus. There are also these things called P bodies that regulate mRNA in the cytoplasm. P bodies are involved in mRNA decay.
Uniparental disomy = the offspring receives 2 copies of a chromosome from 1 parent, and no copy from the other parent; leads to issues with imprinting. Uniparental disomy-> improper imprinting-> Prader-Willi and Angelman syndromes.
Down syndrome is often due to nondisjunction in meoisis, but can also be due to unbalanced Robertsonian translocations (you get too much of one copy of a gene and not enough of the other--this one I took detailed notes about from OnlineMedEd videos in my red notebook; if Robertsonian translocation causes Down syndrome, the pt will actually have a normal number of chromosomes [46], but the amount of genetic information from chromosome 21 will be more than it should be on the chromosomes the pt has, so it presents like there is an extra chromosome 21) or mosaicism (nondisjunction in mitosis causes some cells to have an extra chromosome 21, but not all cells).
Gowers’ sign is when pts with Duchenne muscular dystophy use their arms to get up; looks like they use their arms to “walk up” their own bodies. From Wikipedia:
Gowers' sign is a medical sign that indicates weakness of the proximal muscles, namely those of the lower limb. The sign describes a patient that has to use their hands and arms to "walk" up their own body from a squatting position due to lack of hip and thigh muscle strength.
Duchenne muscular dystrophy is an X-linked recessive mutation in the dystrophin gene. Frameshift and nonsense mutations cause shortened dystrophin gene. In unaffected people, dystrophin links with actin for support of glycoproteins in the plasma membrane of skeletal muscle cells. Defective dystrophin-> breakdown of sarcolemma, degeneration of muscle fibers, calf enlargement (it's really not the calf muscles that are enlarged--it's fat resulting from breakdown of the muscles), increased serum creatine.
Huntington disease is due to CAG trinucleotide repeats in the HTT gene. The more of these there are, the earlier the disease comes on and the more severe it is, which is called anticipation. Huntington's presents with chorea, depression/aggression/apathy, and dementia. Friedreich ataxia, fragile X syndrome, and myotonic dystrophy are also trinucleotide repeat diseases.
From Wikipedia:
Friedreich's ataxia (FRDA or FA) is an autosomal recessive genetic disease that causes difficulty walking, a loss of sensation in the arms and legs and impaired speech that worsens over time. Symptoms generally start between 5 and 20 years of age. Many develop hypertrophic cardiomyopathy and will require a mobility aid such as a cane, walker or wheelchair in their teens. As the disease progresses, people lose their sight and hearing. Other complications include scoliosis and diabetes mellitus.
FRDA is an autosomal recessive disorder that affects a gene (FXN) on chromosome 9 which produces an important protein called frataxin.[5]
In 96% of cases the mutant FXN gene has 90–1,300 GAA trinucleotide repeat expansions in intron 1 of both alleles.[6] This expansion causes epigenetic changes and formation of heterochromatin near the repeat.[5] The length of the shorter GAA repeat is correlated with the age of onset and disease severity.[7] The formation of heterochromatin results in reduced transcription of the gene and low levels of frataxin.[8] People with FDRA might have 5-35% of the frataxin protein compared to healthy individuals. Heterozygous carriers of the mutant FXN gene have 50% lower frataxin levels but this decrease is not enough to cause symptoms.
The condition is caused by mutations in the "FXN" gene on chromosome 9. The FXN gene makes a protein called frataxin. In FRDA, the patient produces less frataxin. Degeneration of nerve tissue in the spinal cord causes the ataxia; particularly affected are the sensory neurons essential for directing muscle movement of the arms and legs through connections with the cerebellum. The spinal cord becomes thinner, and nerve cells lose some myelin sheath.
No effective treatment exists, but there are several therapies in trials. FRDA shortens life expectancy due to heart disease, and some people can live into their sixties or older.
FRDA affects 1 in 50,000 people in the United States and is the most common inherited ataxia. Rates are highest in people of Western European descent. The condition is named after the German physician Nikolaus Friedreich, who first described it in the 1860s.
Homeobox (HOX) genes code for transcription regulators. A homeobox is highly conserved DNA of 180+ nucleotides. Mutations in homeobox genes lead to limbs in the wrong place and skeletal abnormalities. Homeobox genes make sure your leg isn't where your head should be!
From Wikipedia:
A homeobox is a DNA sequence, around 180 base pairs long, found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungi, plants, and numerous single cell eukaryotes.[2] Homeobox genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA to regulate expression of target genes.[3][4][2] Homeodomain proteins regulate gene expression and cell differentiation during early embryonic development, thus mutations in homeobox genes can cause developmental disorders.[5]
Homeosis is a term coined by William Bateson to describe the outright replacement of a discrete body part with another body part, e.g. antennapedia—replacement of the antenna on the head of a fruit fly with legs.[6] The "homeo-" prefix in the words "homeobox" and "homeodomain" stems from this mutational phenotype, which is frequently observed when these genes are mutated in animals. The homeobox domain was first identified in a number of Drosophila homeotic and segmentation proteins, but is now known to be well-conserved in many other animals, including vertebrates.[3][7][8]
CAAT and TATA are promoters necessary to start transcription. CAAT is 75 bases upstreat from the start codon and the TATA (Hogness) box is 25 bases upstream from the start codon. The promoters are where RNA pol II and transcription factors bind.
Heteroplasmy is mixture of two types of genetic material; it's the fact that some cells have normal mitochondria and others have mutated mitochondria because during mitosis, the mutation may distribute more to some cells than others. This affects the severity of the disease. Due to random chance, one offspring may be more severely affected than the other because that's just how the mutation distributed amongst cells during mitosis. This is what happens in syndromes such as MELAS syndrome (Mitochondrial Encephalopyopathy with Lactic Acidosis and Stroke-like episodes), which is due to mutation of mtDNA. All the offspring of an affected mother will be affected because you only get mtDNA from your mom, whose eggs have a lot of it. MELAS syndrome causes seizures, stroke-like episodes, muscle weakness, lactic acidosis.
G6PD deficiency is X-linked recessive; this means that affected males will make unaffected sons (because they can't give their sons the bad X) and carrier daughters (who don't have the disease themselves because they have one good X to counteract the bad X). Females who carry X-linked recessive chromosomes have a 50% change of having affected sons and a 50% chance of having carrier daughters.
An enhancer sequence is found in the introns, upstream, or downstream of a gene. In eukaryotes, RNA pol II makes mRNA from DNA template; enhancer sequences bind to activator proteins that help DNA to bend, which lets the activator proteins interact with the transcription factors and RNA pol II, which causes faster transcription. Silencers bind to repressor proteins and decrease rate of transcription.
Hemophilia A is X-linked recessive = factor VIII deficiency.
Nucleosomes = DNA wrapped around a core of 8 histone proteins. Histone H1 is outside of the histone core of nucleosomes and promotes compaction of heterochromatin.
Ok, I think it's prokaryotes that have DNA pol I, II, and III and eukaryotes that have RNA pol I, II, and III. Eukaryotes have 5 DNA polymerases (alpha, beta, gamma, delta, and epsilon). DNA pol I, II, and III have 3'-> 5' exonuclease (proofreading) capability; but only DNA pol I has 5'-> 3' exonuclease activity, which allows it to remove RNA primers and repair damaged DNA. I got more than one question on this. So remember that DNA pol 1 has 5’-> 3’ exonuclease activity. Eukaryotes have multiple origins of replication whereas prokaryotes have 1.
#wobble#bloom syndrome#uniparental disomy#hnRNA#Down syndrome#Robertsonian translocation#mosaicism#genetics#Duchenne muscular dystophy#heteroplasmy#MELAS#MELAS syndrome#G6PD#activator protein#enhancer sequence#friedreich ataxia#Friedreichs ataxia#HOX#hox gene#homeobox#nucleosome#dna polymerase#DNA pol
4 notes
·
View notes
Text
Assembly & Summary
There is a conserved purine-rich sequence (purines are adenine and guanine) in the central part of the pregenomic RNA (35S RNA) leader sequence. The coat protein of cauliflower mosaic virus interacts with this purine-rich sequence on the leader sequence. We think that the assembly of the double stranded DNA of cauliflower mosaic virus into the viral capsid occurs in inclusion bodies in the cytoplasm. Inclusion bodies are usually aggregates of viral capsid proteins. Thus, the interaction between the pregenomic RNA and the viral protein capsids are likely what brings the viral genome into the inclusion bodies, thus making sure that it ‘floats into the capsid’ before reverse transcription and then gets assembled properly.
Summary of Cauliflower Mosaic Virus (CaMV) Replication Cycle
After introduction into a host cell, the virions migrate to the nuclear envelope where the decapsidate (uncoat). Then the viral genomes are deposited into the nucleus where they form minichromosomes (repairing the nicks supercoils the viral dsDNA and then they bind onto histone complexes). These minichromosomes are transcribed by host RNA Polymerase II which generates two mRNAs. These two are the polycistronic 35S RNA which has the entire genome (encoding 6 proteins), and the monocistronic 19S RNA which encodes a single protein, P6 (TAV, TransActivator Protein).
In the cytoplasm, ribosomes translate the P6 protein from the 19S RNA normally and then the P6 proteins aggregate in small inclusion bodies, which are groups of viral capsid proteins. There, the P6 protein transactivates the translation of all the other viral proteins on the 35S RNA. Then the 35S RNA is additionally used as a template for reverse transcribing (making) viral double stranded DNA (which ultimately gets included into the viral particle as the genome). The reverse transcribed viral dsDNA genome is encapsidated into virions during or shortly after its synthesis. The progeny virions that are generated are most likely grouped together near the inclusion bodies full of P6, and as the inclusion body grows larger it becomes a viral factory.
1 note
·
View note
Note
I apologize in advance for what is pretty much a science lesson (I don't know how much background you have).
Essentially, it may have been the case that no actual DNA mutations occurred. Since he already should possess the Black Arm's genetic information, it would be a case of "switching on" those genes that were previously dormant. This is possible in extant biology through the modification of DNA histones. DNA, when wrapped around these histones are considered "silenced," as when they're tightly packed the proteins that transcribe that genetic information can't get to it to do so. So: tightly packed around histones -> no mRNA (messanger RNA) -> no proteins. What's interesting is that the packing of the DNA around these histones is regulated, meaning that they can be moved around via histone modifications, which can be triggered by environmental factors. In Shadow's case, his chaos control may have happened to cause a histone modification response that resulted in the movement of histones off of his Black Arms genes (and maybe the addition of them to some of his Mobian genes, silencing them), causing the expression of a more significant Black Arms phenotype. Why now? Idk, but maybe a recent life stressor, a different hormonal change, or an external environmental factor may have made the histone modification more liable to occur upon the use of/exposure to chaos energy. If you haven't already heard of this and want to learn more about this concept, I would recommend looking up "epigenetics," it's a fascinating field of study.
Now for the second half: sure, just the expression of some new genes with standard DNA bases would be perfectly sufficient, but I think it'd be fun if there some foreign factors due to the whole "alien race" thing. A fun fact about the DNA nucleobases (the flat rings that hold the genetic information, which they carry primarily through their distinct hydrogen bonding patterns) is that they aren't the only possible ones out there. One example that has been published in more recent years is called the "Hachimoji" (tl: eight character) system (Hachimoji DNA and RNA: A genetic system with eight building blocks, Hoshika et al. 2019 and Visualizing "Alternative Isoinformational Engineered" DNA in A- and B-Forms at High Resolution, Hoshika et al. 2022). With this system, they can add two new sets of synthetic base-pairing nucleobases that can be incorporated into extant DNA (without disrupting the structure too much) and transcribed. Additionally, it would also be fun if some of the proteins that get made from this "ALIEN" DNA were able to synthesize what are for us considered "non-proteinogenic" amino acids and then incorporate them into further new proteins, even though our current set would probably suffice.
In the end, this change would technically be reversible. You've just got to figure out a way to get the histones back to their old spots, so there's hope there.
Hello! I really enjoyed your fic DNA Collision. I'm a bioorganic chemist who technically studies alternative DNA/RNA components (and also knows more than a bit about gene expression). Would you mind if I dumped my personal headcannons as to how Shadow's "mutations" could have occurred from an up-to-date scientific perspective on you?
Omg PLEASE do!! I really love DNA/gene stuff, so I would LOVE to hear everything ;u;
(My reply to your theories will be vague though, since I don't want to spoil my own story, but I am genuinely really curious to hear what you think!)
5 notes
·
View notes
Text
A scalable and cost-efficient rRNA depletion approach to enrich RNAs for molecular biology investigations [Method]
Transcriptomics analyses play pivotal roles in understanding the complex regulatory networks that govern cellular processes. The abundance of rRNAs, which account for 80-90% of total RNA in eukaryotes, limits the detection and investigation of other transcripts. While mRNAs and long non-coding RNAs have polyA(+) tails that are often used for positive selection, investigations of polyA(-) RNAs, such as circular RNAs, histone mRNAs, and small RNAs, typically require the removal of the abundant rRNAs for enrichment. Current approaches to deplete rRNAs for downstream molecular biology investigations are hampered by restrictive RNA input masses and high cost. To address these challenges, we developed rRNA Removal by RNase H (rRRR), a method to efficiently deplete rRNA from a wide range of human, mouse, and rat RNA inputs and qualities at a cost 10-20-fold cheaper than other approaches. We employed probe-based hybridization and enzymatic digestion to selectively target and remove rRNA molecules while preserving the integrity of non-rRNA transcripts. Comparison between rRRR to two commercially available approaches found that they had similar efficiencies at depleting rRNAs and comparable off-target effects. Our developed method provides researchers with a valuable tool for investigating gene expression and regulatory mechanisms across a wide range of biological systems at an affordable price that increases the accessibility for researchers to enter the field, ultimately advancing our understanding of cellular processes. http://rnajournal.cshlp.org/cgi/content/short/rna.079761.123v1?rss=1&utm_source=dlvr.it&utm_medium=tumblr
0 notes