#sub2
Explore tagged Tumblr posts
Text
☕️ ⋆ FIN!

this feels like tj’s worst nightmare coming back to life . because he knows— there’s no coming back from this . no amount of fighting , no desperate pleas , no grand gestures will change the way beau looks at him now . the realization settles deep in his chest , heavier than anything he’s ever carried . as much as he wants to keep fighting for them , for what they used to be , he can see it so clearly now— beau is giving up on him entirely . his heart and mind are set on santi , the one person who is willing to give beau everything , who would never trade him for anything else .
that was supposed to be him .
if he hadn’t been so stupid . if he hadn’t let beau go over a fleeting feeling for nico . if he had only realized sooner what he had before it was too late . but life doesn’t work like that . love doesn’t work like that . one moment , you’re loving someone like they’re the only person meant for you; the next , they’re loving someone else , and you’re just a chapter in their past . it’s cruel how easily people move on , how love shifts and changes when you thought it never would . beau moved on . tj? he doesn’t think he ever will . because if he could love nico as much as he did , then what he feels for beau is bigger than that— something no one else will ever take the place of , even if beau no longer belongs to him .
he understands beau’s wrath , his resentment . that’s why he chooses to let go . he won’t fight anymore . he won’t beg . beau deserves happiness , peace , a love that isn’t weighed down by past mistakes . tj will just have to live with the memories— the way they loved each other when they were young and carefree , too innocent to understand what love really meant . he’ll keep a piece of beau , even from a distance , even in the quiet loneliness of his own mind .
beau’s words blur in his ears , muffled and distorted , as if his body is rejecting them . he doesn’t understand them . doesn’t want to . because to understand means to accept , and he can’t accept that beau wants him to be happy , that hoping he will also find a love that he deserves . he doesn’t want anyone replacing beau . period . if this is his punishment , if this is what he deserves , then so be it . he’ll just live with it . alone .
so , he steps back too .
�� i hope he makes you happy . i hope he gives you the love i failed to give you . ��� his voice is steady , but his heart is breaking all over again . seeing beau cry hurts more than anything . tj can only hope this is the last time he has to see him like this . the last time he has to hurt him .
“ i should go , ” he says quietly , like a final goodbye . “ goodbye , beau . ” and with that , he turns away . walks . never looks back . because if he does , he knows he’ll break . he’ll beg . and he’s already lost him once— he won’t put himself through that again .

the second tj steps into his empty home , he moves like a man possessed . he pulls out his phone , makes the calls he’s been putting off . he contacts his colleagues in the city , tells them he’s accepting the job offer in new york . he doesn’t ask about the details . he just wants to leave . as soon as possible .
that night , he packs everything he owns , shoving clothes into suitcases , throwing away things he doesn’t need . he lists his place on airbnb , a temporary solution for now , but in truth , he knows he’s never coming back . by the time he’s done , the house barely looks lived in . no trace of him remains .
before he leaves for good , he steps outside , pausing in front of the garbage bins . in his hands , he holds a scrapbook— a year’s worth of memories without beau . every single thing he did , every moment he tried to capture , a way to remind himself that life goes on . that he goes on . at the back , a letter he wrote for beau . a long , heartfelt message he was supposed to give him for christmas .
it all feels useless now .
without hesitation , he tosses it into the bin . the scrapbook . the letter . the remnants of a life that no longer belongs to him . he walks away , leaves everything and everyone behind . no goodbyes . no looking back .
because if he does , he might never leave at all .
#wow.......eto ending nila sa sub2. wow......#p!play the ones we once loved#putangina#⠀⠀�� 𝒃𝑒𝑎𝑢 𝖺𝗁𝗇 : general ⁎#⠀⠀𐔌 𝒃𝑒𝑎𝑢 𝖺𝗁𝗇 : prose ⁎#⠀⠀𐔌 𝒗𝑒𝑟𝑠𝑒 : forevermore ⁎#⠀⠀𐔌 𝒇𝑜𝑟𝑒𝑣𝑒𝑟𝗆𝗈𝗋𝖾 : sub² ⁎#⠀⠀𐔌 𝒇𝑜𝑟𝑒𝑣𝑒𝑟𝗆𝗈𝗋𝖾 : sub⁴ ⁎#lovesboat
9 notes
·
View notes
Text
I JUST WANT A MID SHYT TO MAKE OUT WITH AT THE FUCJING CLUBBB IS THAT TOO FAWKING MUCH TO ASK FOR OH NY GODDDDD not sub2 OLD ASS 40 YEAR OLDS GTFOOO😭😭😭😭🙏🙏🙏🙏 im on my KNEES WHERE ARE THE MEN IN THEIR MID 20S YALLL
9 notes
·
View notes
Note
SUBNAUTICA 2 TRAILER CLYDE
I KNOW I SAW!!!!!!!!! Just casually browsing my feed when that lovely alert popped up~
In all seriousness, that teaser makes me really hopeful/hyped for Sub2. I never joined the hate bandwagon for Below Zero, but I found it to just be a solid game, not one that re-captured the magic of the original. This though? This feels like og Subnautica. Even with the arrival of a friend (which might just be a nod to the promised co-op?), the whole teaser has that isolated, alienated feel to it. The world is gorgeous, but every beautiful thing posses a hidden danger. You are just a spec in the deep, dark, terrifying hole that is a foreign ocean. You're alone. The only voice for miles around is the semi-cold AI system trying to keep you alive. You're vulnerable.
There are just so many details here for a minute of info. The loud sound of your breathing. The blinking red alert light. The increasingly frantic beeping. The soft edges as your groggy brain remembers the pretty, safe surface. The fact that the crab-thing appears harmless but can still kill you with its presence alone. The "Oh shit if all the fish are swimming away there's something COMING" callback. The relief of seeing a friend show up before they're snatched by a GIANT BIO-LUMINESCENT CREATURE THAT'S JUST PERSUING ITS NATURAL INSTINCTS BUT ALSO IT'S NATURAL INSTINCTS ARE TERRIFYING." Perfect for spooky season.
(Although I'm ngl, my first thought was that Purple Dude might secretly be a friend like the Emperor Leviathan. It would fit the other parallels of that ending :D)

So yeah I'm PUMPED. Haven't been this excited for a sequel since Hades II, which admittedly hadn't been that long since EA released but STILL. I do have to decide though if I'm going to experience Sub2's EA. A part of me really wants to because a) I missed the initial EA experience and b) I'm impatient, but on the other hand the Subnautica series doesn't lend itself to incomplete play like a rogue-like does and I suspect it will be a lot more bug ridden than Hades II was when it came out... but we'll see. I'll probably join EA if only to avoid spoilers lol
6 notes
·
View notes
Text
OMFG DO YOU THINK SUBNAUTICA THREE WILL BE US GOING BSCK TO CURE THE KARAA ON ITS SOURCE PLANET??
I still haven’t finish sub2 (ik) so maybe I’m forgetting some shit but if I remember correctly didn’t the architects bring the karaa from another planet to their home planet? And the testing to cure it was done on 4546B?
And the lore kind of solves all the loose ends for 4546b and the architects planet so where else do we have to go in this story then the very start of the Karaa? Plus I feel like a lot of people are speculating that karaa is going to be used by Alterra soooo… this is my theories but once again I am stoned and haven’t seen/done a proper play through since sub2 was pre-release.
3 notes
·
View notes
Text
The way I need the new character in Sub2 to be connected to the below zero. Listen give me weird ailen child from below zero. Since it seems like instead of "oh update your machine" it more "update yourself evolution 2".
Its not gonna happen but im actually super excited for subnatica 2!
4 notes
·
View notes
Text
I love to see a Women Win

(Amanda Nunes)
September 19, 2024 Rookie's Playbook
I’m back at it again deep diving into things I have no concept of, and today it’s the world of the UFC! I like for my first read-through something to be an explanation of sorts so I can try and figure out exactly what is happening, and today I'm diving into the beauty of women champions.
From my understanding there are four types of championships, there is feather-weight, bantam-weight, fly-weight, and straw-weight. I’m not gonna lie, half of these to me sound like the same thing with AI generated names. After doing further research it all depends on the weight of the fighter, which when I was younger used to confuse the hell out of me because I thought it was so rude to segregate people into weight categories, now however I know it’s so you don’t get absolutely murdered.
Feather-weight(126 pounds) has no current champion, but they do have three past champions. Amanda Nunes won in 2018 with two defenses and a TKO of 1 over her opponent Cris Cyborg. I’m not going to sit here and lie to you either if my last name was Cyborg no one could talk to me. That is such badass. The only knowledge I have of a TKO(Total Knockout) is when executed the fighter is left unable to complete the fight, so that is insanely hardcore to me. Currently, the brazilian fighter has insane stats ranging from 13-2 TKO, SUB 4-2, and W-L-D 23-5-0. Now, again I will not lie to you I only know what the TKO is.
Sub is when a fighter ‘taps out’, yielding to their opponent so that they win. So, that takes the badass levels down some, but hey who knows this could actually be the lowest for someone and she is the dominant woman to have ever lived. Then the W-L-D is win, loss, or draw. The thing that I'm getting at for Nunes is that she doesn’t believe in draws and eats up her opponent every time, 23 wins! Insane!
Now that I know the terms let's move onto bantamweight, which is the 135 pound limit. The current champion for this category is Raquel Pennington, with an outcome of a UD over Mayra Bueno Silva. Now comes my next question, the hell is a UD? This is a term I have heard of and often yell out when I want to look knowledgeable, it means unanimous decision. This is when all the judges are in agreement of which fighter is the winner, so good for Miss Pennington!
Pennington’s stats are W-L-D 16-9-0, TKO 1-1, and sub is 1-1, not bad but not my new favorite fighter Amanda Nunes good.
Women’s flyweight is next on our list and their poundage (?) is 125. I’m going to be honest, with all the searching and rules about the fighters weight is insane to me, what is a one pound difference in the grand scheme of it all you know? Anyway, the current champion of this category is Valentina Shevchenko who won in a unanimous decision against Alexas Grasso! Look at me knowing my terms, this is what we call development guys. Shevchenko’s stats are W-L-D 24-4-1, TKO 8-1, and Sub 7-1. I’m gonna be honest, Miss Nunes has my heart, but Schevchenko scares the shit outta me the most.
The final category is straw-weight and the weight is 115 pounds, again the names just all seem too similar for my taste and I find them very much confusing but non the less I digress. The current champion is the lady with the best cheekbones I have ever seen Zhang Weili who won against Carla Esparza with a Sub2. Her stats rival Nunes to me with a W-L-D 25-3-0, TKO 11-1, and Sub 8-0. She is a Chinese fighter whose career I feel I will be following closely.
After all that research I can confirm that this is the most intriguing of the sports I have covered so far, and I simply cannot wait to learn more about it.
2 notes
·
View notes
Text
okay sub legacy thoughts w spoilers (by 'spoilers' i mean comments on changes made from the OG not plot spoilers) (i am a little way into the lab)
keeping the coin but moving the code to get the fuse elsewhere? DICK move. took me ages to find it. not sure if it's randomised but if it is then i think i got unlucky with the second 2 digits being 11 bcos it wasn't actually obvious that they were numbers when i found them
oughhhhh took me till sub2 to recognise the dimensional co-ordinates i was running into that's really neat and im so hype to see where we're going with that
where is the spoon and the fork... where is my classic submachine cutlery puzzle :(
i appreciate that there's more explanation for what you're doing w the chemicals in the lab (that's always been a bit of a ?? puzzle as it seems to hinge on knowledge that the player doesn't actually have so you're basically just combining items till you hit on the right combo). however i always liked the detail that you're straight up breaking in rather than hunting around for the code for entry so i wish that had been kept!!
!!!!! when re-opening the game and spotting sub 0 added to the level map
ok i need to stop now bcos it's late and also im supposed to be reducing screen time bcos of my eyes. off to bed shortly zzz good night.
14 notes
·
View notes
Text

TWIN PEAKS
s01 - sUB
02 - sUB1 / sUB2
Fuego Camina Conmigo (1992)
03 - sUB
7 notes
·
View notes
Text
i remember like. literally two years ago talking to someone about how sub2:30 seemed almost impossible for AA and now fein has a sub2:15. that is fucking insane but also if anyone could do it it would be feinberg.
#sorry i have mcsrtism i just dont talk about it very much these days#vodwatching his 2:14 rn because i didnt see it live. and i am just so in awe of his talents
3 notes
·
View notes
Text
it is a good day bc i got a sub2 time in set yayyyyyyy
7 notes
·
View notes
Text
#Tutorial: Switch Menu
Nesse tutorial você vai aprender a montar o Switch Menu para seu Tumblr, Blog e Afins.
Pra quem não conhece, para aparecer o conteúdo, basta clicar no assunto desejado.

Primeiro passo - Antes de </head> Cole isso aqui lá.
Segundo passo - Crie os botões no seu CSS, do jeito que você quiser, vou colocar o meu, se usar, credite ok? não custa nada!
/**Menu - Endstonight|Tumblr**/ .menutitle{color: #387377; font-family: ‘Yanone Kaffeesatz’, georgia; font-size:16px; line-height:28px; font-weight:normal; padding:5px; margin-bottom:8px; text-align: left; letter-spacing:1px; background: #f0f0f0;text-indent : 10px;-webkit-transition-duration: .40s;} .menutitle:hover{text-indent : 19px;}
( PS: Link da Font - <link href='http://fonts.googleapis.com/css?family=Yanone+Kaffeesatz’ rel='stylesheet’ type='text/css’>
cole antes de </head> )
Terceiro Passo - Faça o iframe comum no seu menu, e depois monte ele, mais ou menos assim:
<div id=“tutos” style=“display:none”>
<div class=“box”><h2>Tutoriais</h2>
<div id=“topdiv”>
<div class=“menutitle” onclick=“SwitchMenu('sub1’)” style=“cursor:pointer” >Menus</div>
<span class=“submenu” id=“sub1”>
Conteudoo
</span>
Obs: Se você for fazer outros menus, basta substituir o sub1 por "sub2" , "sub3" , "sub4", você pode fazer quantos quiser.
Obs 2: Não retire a ID “topdiv” não vai funcionais sem ela.
0 notes
Text
I am looking for a tutor workers Who can handle simple Typing job https://www.gn3atrk.com/2DB368NW/36M98FTS/?sub2=1.2

0 notes
Text
WK4 - Testing a Potential Moderator
For the this example I used parte of the same code used um Wk3, ralatered to Salaries. It was used Salary as the moderator between years of experience and Age.
The rest of the Code:
Define income groups correctly
def Income(sal): if sal['Salary'] <= 10000:
return 1
elif sal['Salary'] <= 50000:
return 2
elif sal['Salary'] <= 100000:
return 3
elif sal['Salary'] <= 150000:
return 4
else: # This ensures salaries > 200000 are assigned group 3 return 5
data_clean['Income'] = data_clean.apply(lambda sal: Income(sal), axis=1)
Create subsets
sub1 = data_clean[data_clean['Income'] == 1] sub2 = data_clean[data_clean['Income'] == 2] sub3 = data_clean[data_clean['Income'] == 3] sub4 = data_clean[data_clean['Income'] == 4] sub5 = data_clean[data_clean['Income'] == 5]
Compute correlation for each subset separately
if len(sub1) > 1: print('Association between Age and Salary for Very Low Income Salary:') print(scipy.stats.pearsonr(sub1['Age'], sub1['Salary']))
if len(sub2) > 1: print('\nAssociation between Age and Salary for Low Income Salary:') print(scipy.stats.pearsonr(sub2['Age'], sub2['Salary']))
if len(sub3) > 1: print('\nAssociation between Age and Salary for Medium Income Salary:') print(scipy.stats.pearsonr(sub3['Age'], sub3['Salary']))
if len(sub4) > 1: print('\nAssociation between Age and Salary for High Income Salary:') print(scipy.stats.pearsonr(sub4['Age'], sub4['Salary']))
if len(sub5) > 1: print('\nAssociation between Age and Salary for Very High Income Salary:') print(scipy.stats.pearsonr(sub5['Age'], sub5['Salary']))
Bar Chart: Average Salary by Income Group
income_salary_avg = data_clean.groupby('Income')['Salary'].mean()
plt.figure(figsize=(8, 5)) sns.barplot(x=income_salary_avg.index, y=income_salary_avg.values, palette="viridis") plt.xlabel("Income Group") plt.ylabel("Average Salary") plt.title("Average Salary by Income Group") plt.xticks(ticks=range(0, 5), labels=["Very Low", "Low", "Medium", "High", "Very High"]) plt.show()

Scatter plot for Age vs Salary by Income Group
plt.figure(figsize=(8, 5)) # Set figure size sns.scatterplot( x=data_clean["Age"], y=data_clean["Salary"], hue=data_clean["Income"], # Color by Income Group palette="viridis", # Choose a color palette alpha=0.7 # Adjust transparency for better visualization ) plt.xlabel("Years of Experience") plt.ylabel("Salary") plt.title(" Age vs Salary by Income Group") plt.show()
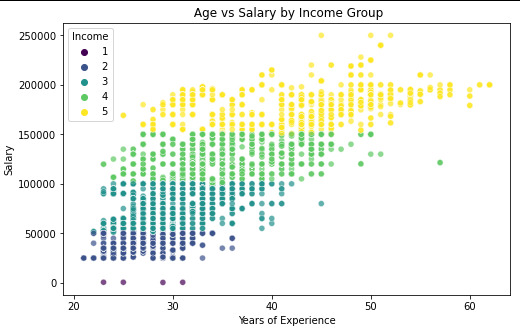
Scatter plot for Years of Experience vs Salary by Income Group
plt.figure(figsize=(8, 5)) sns.scatterplot( x=data_clean["Years of Experience"], y=data_clean["Salary"], hue=data_clean["Income"], palette="coolwarm", alpha=0.7 ) plt.xlabel("Years of Experience") plt.ylabel("Salary") plt.title("Experience vs Salary by Income Group") plt.show()
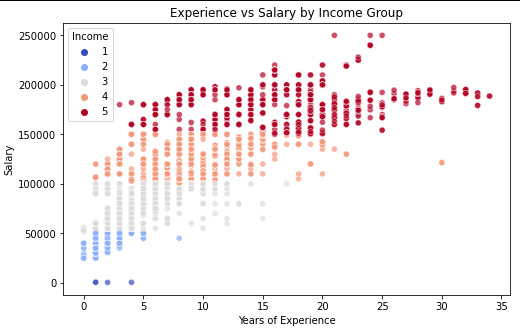
Scatter plot for Age vs Salary with Regression line styling
plt.figure(figsize=(8, 5))
sns.regplot( x=data_clean["Age"], y=data_clean["Salary"], scatter_kws={"alpha": 0.5, "color": "blue"}, # Scatter points styling line_kws={"color": "red"}, # Regression line styling ) plt.xlabel("Age") plt.ylabel("Salary") plt.title("Regression Plot: Age vs Salary") plt.show()

Scatter plot for Years of Experience vs Salary
sns.regplot( x=data_clean["Years of Experience"], y=data_clean["Salary"], scatter_kws={"alpha": 0.5, "color": "green"}, line_kws={"color": "red"}, ) plt.xlabel("Years of Experience") plt.ylabel("Salary") plt.title("Regression Plot: Years of Experience vs Salary") plt.show()

Scatter plot for Age vs Salary by Income Group
plt.figure(figsize=(8, 5))
sns.lmplot( x="Age", y="Salary", hue="Income", data=data_clean, palette="viridis", height=5, aspect=1.2 ) plt.xlabel("Age") plt.ylabel("Salary") plt.title("Regression Plot: Age vs Salary by Income Group") plt.show()
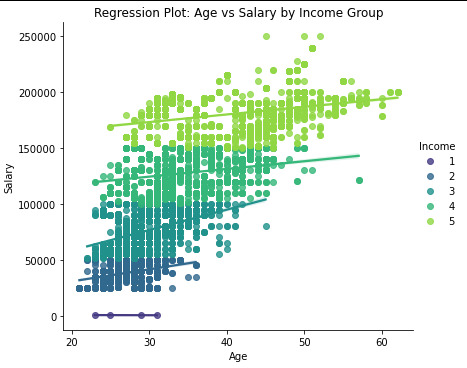
Result:

It is possible to verify that correlation between the Age and salary is for Very Low income, is -0.64, which means it ia a negative correlation and has a p-value=0.35, that is over thant p-value<0.05. so p-value for the very low it is no significante.
for the rest of the groups, Low, Medium, High and Very High, the correlation value is positive, and the p-value is unde 0.05, so we cansay that p-valeu for the rest is a significate
it is possivel to see in the Scatter plot .
0 notes
Text
The Ultimate Guide to Buying Real Estate Using Creative Financing| William Tingle
Private Money Academy Conference:
Free Report:
William Tingle, Host of www.Sub2Deals.com is a nationally known real estate investor, investing coach, author, and public speaker. William had worked in the restaurant business for almost 20 years when in 1999, he ordered a Carleton Sheets Nothing Down course off of a late-night TV infomercial. He read it and took a $5000 advance from a credit card to start his real estate investing career.
Exactly one year later he quit his job for good, paid the credit card off, and has to this day never used a penny of his own money for investing.
For over 10 years he operated a real estate business where he wholesaled and rehabbed numerous properties each year but found his real niche in what he calls “Sub2”, buying subject to existing financing. To date, he has taken the deed on over 500 properties and even though he “retired” in Belize in 2010, he continues to buy 20 to 25 properties a year in this manner in select markets throughout the United States.
He has trained and coached countless students all over the country to become financially successful real estate investors. William has written several real estate courses. His flagship courses are “The “Ultimate” Sub2 Guidebook” & “Extreme Marketing – The “Ultimate” Marketing Guidebook”. “Ultimate Sub2” covers every aspect of “subject to” investing from marketing to finding motivated sellers, to negotiating the deal, to completing the paperwork, to how to market and sell the properties.
“Extreme Marketing” is a marketing manual with literally hundreds of both time-tested and unique, outside-the-box marketing ideas to grow your real estate business.
Timestamps:
00:01 Raising Private Without Asking For It
06:15 Struggling with Mortgage Payments
07:20 Property Sale with Retained Mortgage
11:46 Swift Real Estate Solutions Offered
16:13 Cash Buying Criteria Explained
18:24 Profitable Low-Equity House Investments
22:40 The Rise of Due on Sale Clauses
26:09 Private Lending and Second Position Strategy
27:05 Creative Financing with Seller Partnerships
30:06 Successful Real Estate Strategy

Have you read Jay’s new book: Where to Get The Money Now?
It is available FREE (all you pay is the shipping and handling) at
What is Private Money? Real Estate Investing with Jay Conner
Jay Conner is a proven real estate investment leader. He maximizes creative methods to buy and sell properties with profits averaging $67,000 per deal without using his own money or credit.
What is Real Estate Investing? Live Private Money Academy Conference
youtube
YouTube Channel
Apple Podcasts:
Facebook:
#youtube#flipping houses#private money#real estate#jay conner#real estate investing#raising private money#real estate investing for beginners#foreclosures#passive income
0 notes
Text
Data Visualization - Assignment 2
For my first Python program, please see below:
-----------------------------PROGRAM--------------------------------
-- coding: utf-8 --
""" Created: 27FEB2025
@author:Nicole taylor """ import pandas import numpy
any additional libraries would be imported here
data = pandas.read_csv('GapFinder- _PDS.csv', low_memory=False)
print (len(data)) #number of observations (rows) print (len(data.columns)) # number of variables (columns)
checking the format of your variables
data['ref_area.label'].dtype
setting variables you will be working with to numeric
data['obs_value'] = pandas.to_numeric(data['obs_value'])
counts and percentages (i.e. frequency distributions) for each variable
Value1 = data['obs_value'].value_counts(sort=False) print (Value1)
PValue1 = data['obs_value'].value_counts(sort=False, normalize=True) print (PValue1)
Gender = data['sex.label'].value_counts(sort=False) print (Gender)
PGender = data['sex.label'].value_counts(sort=False, normalize=True) print (PGender)
Country = data['Country.label'].value_counts(sort=False) print (Country)
PCountry = data['Country.label'].value_counts(sort=False, normalize=True) print (PCountry)
MStatus = data['MaritalStatus.label'].value_counts(sort=False) print (MStatus)
PMStatus = data['MaritalStatus.label'].value_counts(sort=False, normalize=True) print (PMStatus)
LMarket = data['LaborMarket.label'].value_counts(sort=False) print (LMarket)
PLMarket = data['LaborMarket.label'].value_counts(sort=False, normalize=True) print (PLMarket)
Year = data['Year.label'].value_counts(sort=False) print (Year)
PYear = data['Year.label'].value_counts(sort=False, normalize=True) print (PYear)
ADDING TITLES
print ('counts for Value') Value1 = data['obs_value'].value_counts(sort=False) print (Value1)
print (len(data['TAB12MDX'])) #number of observations (rows)
print ('percentages for Value') p1 = data['obs_value'].value_counts(sort=False, normalize=True) print (PValue1)
print ('counts for Gender') Gender = data['sex.label'].value_counts(sort=False) print(Gender)
print ('percentages for Gender') p2 = data['sex.label'].value_counts(sort=False, normalize=True) print (PGender)
print ('counts for Country') Country = data['Country.label'].value_counts(sort=False, dropna=False) print(Country)
print ('percentages for Country') PCountry = data['Country.label'].value_counts(sort=False, normalize=True) print (PCountry)
print ('counts for Marital Status') MStatus = data['MaritalStatus.label'].value_counts(sort=False, dropna=False) print(MStatus)
print ('percentages for Marital Status') PMStatus = data['MaritalStatus.label'].value_counts(sort=False, dropna=False, normalize=True) print (PMStatus)
print ('counts for Labor Market') LMarket = data['LaborMarket.label'].value_counts(sort=False, dropna=False) print(LMarket)
print ('percentages for Labor Market') PLMarket = data['LaborMarket.label'].value_counts(sort=False, dropna=False, normalize=True) print (PLMarket)
print ('counts for Year') Year = data['Year.label'].value_counts(sort=False, dropna=False) print(Year)
print ('percentages for Year') PYear = data['Year.label'].value_counts(sort=False, dropna=False, normalize=True) print (PYear)
---------------------------------------------------------------------------
frequency distributions using the 'bygroup' function
print ('Frequency of Countries') TCountry = data.groupby('Country.label').size() print(TCountry)
subset data to young adults age 18 to 25 who have smoked in the past 12 months
sub1=data[(data[TCountry]='United States of America') & (data[Gender]='Sex:Female')]
make a copy of my new subsetted data
sub2 = sub1.copy()
frequency distributions on new sub2 data frame
print('counts for Country')
c5 = sub1['Country.label'].value_counts(sort=False)
print(c5)
upper-case all DataFrame column names - place afer code for loading data aboave
data.columns = list(map(str.upper, data.columns))
bug fix for display formats to avoid run time errors - put after code for loading data above
pandas.set_option('display.float_format', lambda x:'%f'%x)
-----------------------------RESULTS--------------------------------
runcell(0, 'C:/Users/oen8fh/.spyder-py3/temp.py') 85856 12 16372.131 1 7679.474 1 462.411 1 8230.246 1 4758.121 1 .. 1609.572 1 95.113 1 372.197 1 8.814 1 3.796 1 Name: obs_value, Length: 79983, dtype: int64
16372.131 0.000012 7679.474 0.000012 462.411 0.000012 8230.246 0.000012 4758.121 0.000012
1609.572 0.000012 95.113 0.000012 372.197 0.000012 8.814 0.000012 3.796 0.000012 Name: obs_value, Length: 79983, dtype: float64
Sex: Total 28595 Sex: Male 28592 Sex: Female 28595 Sex: Other 74 Name: sex.label, dtype: int64 Sex: Total 0.333058 Sex: Male 0.333023 Sex: Female 0.333058 Sex: Other 0.000862 Name: sex.label, dtype: float64 Afghanistan 210 Angola 291 Albania 696 Argentina 768 Armenia 627
Samoa 117 Yemen 36 South Africa 993 Zambia 306 Zimbabwe 261 Name: Country.label, Length: 169, dtype: int64
Afghanistan 0.002446 Angola 0.003389 Albania 0.008107 Argentina 0.008945 Armenia 0.007303
Samoa 0.001363 Yemen 0.000419 South Africa 0.011566 Zambia 0.003564 Zimbabwe 0.003040 Name: Country.label, Length: 169, dtype: float64
Marital status (Aggregate): Total 26164 Marital status (Aggregate): Single / Widowed / Divorced 26161 Marital status (Aggregate): Married / Union / Cohabiting 26156 Marital status (Aggregate): Not elsewhere classified 7375 Name: MaritalStatus.label, dtype: int64
Marital status (Aggregate): Total 0.304743 Marital status (Aggregate): Single / Widowed / Divorced 0.304708 Marital status (Aggregate): Married / Union / Cohabiting 0.304650 Marital status (Aggregate): Not elsewhere classified 0.085900 Name: MaritalStatus.label, dtype: float64
Labour market status: Total 21870 Labour market status: Employed 21540 Labour market status: Unemployed 20736 Labour market status: Outside the labour force 21710 Name: LaborMarket.label, dtype: int64
Labour market status: Total 0.254729 Labour market status: Employed 0.250885 Labour market status: Unemployed 0.241521 Labour market status: Outside the labour force 0.252865 Name: LaborMarket.label, dtype: float64
2021 3186 2020 3641 2017 4125 2014 4014 2012 3654 2022 3311 2019 4552 2011 3522 2009 3054 2004 2085 2023 2424 2018 3872 2016 3843 2015 3654 2013 3531 2010 3416 2008 2688 2007 2613 2005 2673 2002 1935 2006 2700 2024 372 2003 2064 2001 1943 2000 1887 1999 1416 1998 1224 1997 1086 1996 999 1995 810 1994 714 1993 543 1992 537 1991 594 1990 498 1989 462 1988 423 1987 423 1986 357 1985 315 1984 273 1983 309 1982 36 1980 36 1970 42 Name: Year.label, dtype: int64
2021 0.037109 2020 0.042408 2017 0.048046 2014 0.046753 2012 0.042560 2022 0.038565 2019 0.053019 2011 0.041022 2009 0.035571 2004 0.024285 2023 0.028233 2018 0.045099 2016 0.044761 2015 0.042560 2013 0.041127 2010 0.039788 2008 0.031308 2007 0.030435 2005 0.031134 2002 0.022538 2006 0.031448 2024 0.004333 2003 0.024040 2001 0.022631 2000 0.021979 1999 0.016493 1998 0.014256 1997 0.012649 1996 0.011636 1995 0.009434 1994 0.008316 1993 0.006325 1992 0.006255 1991 0.006919 1990 0.005800 1989 0.005381 1988 0.004927 1987 0.004927 1986 0.004158 1985 0.003669 1984 0.003180 1983 0.003599 1982 0.000419 1980 0.000419 1970 0.000489 Name: Year.label, dtype: float64
counts for Value 16372.131 1 7679.474 1 462.411 1 8230.246 1 4758.121 1 .. 1609.572 1 95.113 1 372.197 1 8.814 1 3.796 1 Name: obs_value, Length: 79983, dtype: int64
percentages for Value 16372.131 0.000012 7679.474 0.000012 462.411 0.000012 8230.246 0.000012 4758.121 0.000012
1609.572 0.000012 95.113 0.000012 372.197 0.000012 8.814 0.000012 3.796 0.000012 Name: obs_value, Length: 79983, dtype: float64
counts for Gender Sex: Total 28595 Sex: Male 28592 Sex: Female 28595 Sex: Other 74 Name: sex.label, dtype: int64
percentages for Gender Sex: Total 0.333058 Sex: Male 0.333023 Sex: Female 0.333058 Sex: Other 0.000862 Name: sex.label, dtype: float64
counts for Country Afghanistan 210 Angola 291 Albania 696 Argentina 768 Armenia 627
Samoa 117 Yemen 36 South Africa 993 Zambia 306 Zimbabwe 261 Name: Country.label, Length: 169, dtype: int64
percentages for Country Afghanistan 0.002446 Angola 0.003389 Albania 0.008107 Argentina 0.008945 Armenia 0.007303
Samoa 0.001363 Yemen 0.000419 South Africa 0.011566 Zambia 0.003564 Zimbabwe 0.003040 Name: Country.label, Length: 169, dtype: float64
counts for Marital Status Marital status (Aggregate): Total 26164 Marital status (Aggregate): Single / Widowed / Divorced 26161 Marital status (Aggregate): Married / Union / Cohabiting 26156 Marital status (Aggregate): Not elsewhere classified 7375 Name: MaritalStatus.label, dtype: int64
percentages for Marital Status Marital status (Aggregate): Total 0.304743 Marital status (Aggregate): Single / Widowed / Divorced 0.304708 Marital status (Aggregate): Married / Union / Cohabiting 0.304650 Marital status (Aggregate): Not elsewhere classified 0.085900 Name: MaritalStatus.label, dtype: float64
counts for Labor Market Labour market status: Total 21870 Labour market status: Employed 21540 Labour market status: Unemployed 20736 Labour market status: Outside the labour force 21710 Name: LaborMarket.label, dtype: int64
percentages for Labor Market Labour market status: Total 0.254729 Labour market status: Employed 0.250885 Labour market status: Unemployed 0.241521 Labour market status: Outside the labour force 0.252865 Name: LaborMarket.label, dtype: float64
counts for Year 2021 3186 2020 3641 2017 4125 2014 4014 2012 3654 2022 3311 2019 4552 2011 3522 2009 3054 2004 2085 2023 2424 2018 3872 2016 3843 2015 3654 2013 3531 2010 3416 2008 2688 2007 2613 2005 2673 2002 1935 2006 2700 2024 372 2003 2064 2001 1943 2000 1887 1999 1416 1998 1224 1997 1086 1996 999 1995 810 1994 714 1993 543 1992 537 1991 594 1990 498 1989 462 1988 423 1987 423 1986 357 1985 315 1984 273 1983 309 1982 36 1980 36 1970 42 Name: Year.label, dtype: int64
percentages for Year 2021 0.037109 2020 0.042408 2017 0.048046 2014 0.046753 2012 0.042560 2022 0.038565 2019 0.053019 2011 0.041022 2009 0.035571 2004 0.024285 2023 0.028233 2018 0.045099 2016 0.044761 2015 0.042560 2013 0.041127 2010 0.039788 2008 0.031308 2007 0.030435 2005 0.031134 2002 0.022538 2006 0.031448 2024 0.004333 2003 0.024040 2001 0.022631 2000 0.021979 1999 0.016493 1998 0.014256 1997 0.012649 1996 0.011636 1995 0.009434 1994 0.008316 1993 0.006325 1992 0.006255 1991 0.006919 1990 0.005800 1989 0.005381 1988 0.004927 1987 0.004927 1986 0.004158 1985 0.003669 1984 0.003180 1983 0.003599 1982 0.000419 1980 0.000419 1970 0.000489 Name: Year.label, dtype: float64
Frequency of Countries Country.label Afghanistan 210 Albania 696 Angola 291 Antigua and Barbuda 87 Argentina 768
Viet Nam 639 Wallis and Futuna 117 Yemen 36 Zambia 306 Zimbabwe 261 Length: 169, dtype: int64
-----------------------------REVIEW--------------------------------
So as you can see in the results. I was able to show the Year, Marital Status, Labor Market, and the Country. I'm still a little hazy on the sub1, but overall it was a good basic start.
0 notes