#my lab environment is good my pi is understanding
Explore tagged Tumblr posts
Text
1d. First Day at the Lab - Outliers
Name: Ren
Day: 1
Funds: $ 100
Today is my first day working at Ar Leith Labs - I can't believe I finally landed a job!
To be honest, I didn't look too deeply into what they do at Ar Leith Labs - I basically sent my curriculum to every neuroscience research lab that was hiring. Now that I'm here, I can't even find a pamphlet explaining the research in detail.
Ok, I'll be professional and go introduce myself to my coworkers now; either them or the PI can tell me more about the job.
---
There are only two other research assistants in my group: Leanne and Perry; neither of them seems to be the chatty type, at least not with me. I was looking forward to meeting my PI, but Leanne told me that she has never shown her face around here.
Right then we heard the PI speak; it felt as if she was standing right next to us. This lab must have a pretty technologically advanced speaker system!
The PI's voice welcomed me and introduced herself as C.N, just her initials; she invited me to get acquainted with the lab environment, and help my coworkers out with anything they might need.
I found it a little odd that she's not meeting us in person, but maybe there's an excellent reason for it. I don't want to pry, especially not on my first day.
Nobody was available to give me a tour - lots of work to do, which is fair - so I walked around the lab by myself, studying the equipment. I didn't recognize any of the machines, except for the obvious desktop computer in the corner. That one even looks a little old, in contrast with the rest of the devices.
Leanne noticed me looking at the computer and asked if I know how to code; heck yeah I do, I took a few classes and I'm pretty ok at it! So she asked me to write a bit of code to generate graph data for her latest research data. It's strange that they don't have software for that already, but I decided to avoid asking any questions.
I took this opportunity to look over the data, hoping it would clarify what kind of research we're supposed to do here, but I couldn't make heads or tails of it. Oh, well.
I couldn't recognize the OS the computer is running either, but it seemed loosely based on Unix. I was making good progress until I started testing my code; I got the error "Unable to find or open '/Brain/TemporalLobe/Hippocampus/MISTAKE.png'". I'm 100% sure I never referenced this file, and what a bizarre name!
I immediately thought this must be a prank - Leanne and Perry must have planned this as a funny welcome for me. I resolved to laugh and tell them it was a cool prank; that would show them I'm chill.
Unfortunately, they kept insisting they didn't know what I was talking about. They looked annoyed, so I assume they were being truthful. Alright, time to debug.
A quick search of the codebase and external libraries for the file path in the error message yielded no results. I tried looking just for "Mistake.png" and got nothing once again.
Interestingly enough, though, "Ren/Brain/" exists, except there's only a "temp" folder in there. Maybe I don't have the right access levels to see other folders? There doesn't seem to be a root user either.
I bothered Leanne and Perry to see whether they have access to the other folders - they don't, but they have their own users on this machine, with their own "/Brain/" folders. Also, my code wasn't available to them. They said the users were already set up for them when they joined, just like mine; IT support must be incredible around here.
In the end I decided to share the code with Leanne's user, in the off chance it would work for her. It did, just like I hoped, and Leanne got her graph.
I don't fully understand, but... great. Maybe I should talk to the IT support people, or take a few more coding classes.
---
The rest of the day was spent on boring menial tasks.
I bet my coworkers think I'm more trouble than I'm helpful, but hey - they'll change their minds, soon enough. After all, I didn't graduate top of my class just to be ignored at my job.
Luckily, at least C.N. already sees potential in me: before I left for the day, she said tomorrow I'll be tabling at an event called "The Gathering"! My first table, and on day 2? I can't wait!
I forgot to ask for the address, but I bet I can find all the info I need online. I'm obviously being tested, and I will show initiative, dependability, and bring a ton of new participants for the study!
--------------------
This is a playthrough of a solo TTRPG called "Outliers", by Sam Leigh, @goblinmixtape.
You can check it out on itch.io: https://far-horizons-co-op.itch.io/outliers
#indie ttrpg#itch.io#journal entry#journaling#playthrough#ttrpg#solo games#solo ttrpg#tabletop role playing game#tabletop roleplaying#outliers#lab notes#outliers ttrpg
12 notes
·
View notes
Note
Hi! I'm honestly excited that tumblr recommended me a good blog for once. It's nice to see other trans ppl living their best life out here.
Anyway, I hope you don't mind a question, but I'm currently an undergrad and really interested in grad school. idk how applicable your experience would be since im a comp sci student, but what's it like in grad school? how accepting has it been of trans people for you? do you have any recommendations on what to do to increase the chance of getting in?
p.s. idk a ton about biochemistry or genetics, but i have a friend who's super into it (he's also planning on going to grad school for it lol), and I love hearing him talk about it. so I'm also curious as to what you're doing :o
Grad school is broken up into phases in my experience, which is dependent on institution and major. For me, year one is classes and rotations, the latter of which is 7 week trials in a lab of interest to get a feel for their work and the lab social and mentor environ. Classes generally compose discussing research techniques and recent publications, and test you on your ability to read research literature and understand it as well as propose follow up investigations. Year two is finishing classes and starting work on your thesis projects, as well as completing preliminary exams, which for biochem are written (grant proposal) and oral (ppt presentation of grant proposal). Year two has less classes than year one and your actual lab work dominates most of your time. Year 3, 4, and 5 are basically spent having a full time job as a senior lab researcher where you are expected to learn your local field well enough to come up with an idea and pursue it to discover or demonstrate something novel in the form of your thesis, as well as publish at least one paper on your work in the meantime.
My institution has not had any issues with trans ppl in my experience. I came out to my lab in Oct of 2023, and gradually to my classmates and the wider program over the following months. Administrative staff and professors have been very on the ball with my pronouns and chosen name, and our program recently got a new professor who is also a trans woman. I won't say which institution to reduce the amount i dox myself, but safe to say I'm content in regards to my treatment as a transgender person.
Getting into grad school usually requires demonstrated undergraduate research or a gap period of research employment after undergrad. I did some undergraduate work (though not strictly in my field) and a year of relevant employment after undergrad. 3 months into said employment I applied for grad school lol which was a little earlier than usually advised bc i hadn't been there long. (My post-bac PI was very demanding and restrictive so I wanted out ASAP). The main things grad schools are looking for is are you able to talk about research you're doing now, what research you want to do, and why you've chosen their institution. They want you to demonstrate you have an interest in researching topics they have professors willing to support, and also that you have the problem solving and data analysis skills to adequately perform research work and operate semi-independently. Having undergraduate or post-bac research that you can explain in detail why you did certain things, what the motivation is, what the goal is, etc is how you prove your worth to grad recruiters (who are usually professors looking for students).
My work is specifically focused on the use of short synthetic peptides to sense the activity of kinases, which regulate pathways in the human body related to growth, division, and apoptosis. When kinases are disregulated it causes various diseases, but it's hard to test drugs for many of these kinases bc there aren't effective monitors for their activity that don't get a lot of false positives due to overlapping signal pathways. The use of synthetic peptides with certain sequences gets around that issue of false positives, and they can be modified to provide information about how effectively a kinase is binding and interacting with the peptide, which would change under exposure to effective inhibitor drugs for those kinases. TL;DR I make little bits of protein to bind other broken proteins and tell me when they're turned on so other ppl can design drugs to turn them off.
Pls let me know if you have any further questions, I apologize that I don't know any comp-sci majors so this might be wildly inaccurate in some regards. XD
11 notes
·
View notes
Note
I'm searching for a gap year job and I'm looking at a lot of lab tech positions. The lab I'm in now has 6 full time researchers and a few undergrads, but the lab I'm looking to join has about 40 people total and I'm kind of nervous about that big of a switch. Are there benefits to working in a large lab?
Hello Anon!
During my gap year(s) I also worked as a lab tech in a medium-ish sized lab (13 people), that was then part of a close-knit department comprised of 40+ people. We regularly worked with everything, including going to all the meetings, sharing equipment, collaborating on experiments, etc. It was like being in a lab inside a lab! Here are my own pros vs cons of working in a huge lab:
Pros:
A history of success. A lab doesn't become that large without a constant flow of grant money, and a constant flow of grant money usually means a regular churning out of high-quality publications from high-quality research.
Lots of resources: be it mentors, equipment, lab space (hopefully), money (presumably), internal and external collaborators, and sources of help. There was never a moment where I couldn’t find at least one person to help me out with something. A note about mentors: a large lab will usually have Scientists (with PhD's) who head their own projects. So the chain of supervising would go to them first, then the PI.
More variety of cool research to do and learn about. This is great if you have a passion for learning new techniques, and will also broaden your CV. In addition, if you help out with various projects, you have the opportunity to be a co-author on all those papers.
Usually will have more dedicated positions, like a lab manager whose full time job is only admin-related stuff, or someone who cleans all the glassware/makes cell media/etc. It takes the multi-tasking away from actual research.
Really organized. You just have to be in an environment like this (but on the downside, if someone deviates from the organization system in place, it could cause more chaos than in a smaller lab with less material).
Company at all times. There will always be someone around, so that's great if you don't like to ever be alone in the lab.
More redundancy in expertise. One downside of a small lab is that if one members is out, their project would come to a grinding halt (or they’ll be bombarded with inquires about where things are and how to do this and that--super frustrating). But in larger labs, the other members can help out.
Your lab will probably be well known, even outside of the institution, so that's useful in leveraging that "brand" during the rest of your career.
Related, you will have a large network of lab alumni who can also help you in your next career steps.
More variety of fun outside-of-lab activities. The large lab I was a part of had amazing potlucks (sooo much food) and even their own soccer games on the weekend.
Cons:
Can be overwhelming, even months in. The learning curve will be higher for fully understanding all the different projects happening at once.
PI may be spread thin, and may not give you the one-on-one time you want (but that can be remedied by the presence of other mentors in the lab).
Can be loud and slightly chaotic at times, which just comes with the territory of being around more people. There may also be a higher chance of interpersonal friction between lab members.
More people will be using the same equipment, so planning and communication are crucial to properly share them.
Personal work space (like your own desk or lab bench) may be limited. Hopefully the lab isn't hiring more people than it can fit, but if having your own space is important to you, be sure to inquire about that during the interview.
Specifically for Covid-19: Social distancing may be harder, and there may be personnel scheduling “rotations” in place (like spreading out shifts during the day, which may involve some members coming in during the evenings/weekends). This is something to definitely bring up in your interviews.
That’s all I can think about from the top of my head. If anyone else has experience working in large labs, feel free to chime in!
Good luck anon with your job hunt! I hope it all works out.
14 notes
·
View notes
Note
Have you thought about what kind of profs Cedric Junpier, Fennel, and Ivy would be? :D
!! omg people really liked my pokemon professor post huh... i'm flattered :'D guess i'm a pokemon blog now (jk)
professor mister juniper: i think he's retired now and he's so so proud of how much his little girl has grown up into a professor like him :'0 enthusiasm for ecology/evolution and a passion for spreading it around runs in the family. he was also a successful professor (at the same university his daughter teaches at now!) who did lots of great stuff in biogeography and ecology and fired up lots of future scientists. he seems to travel a lot and give talks occasionally, and will make funny cameos in selfies on aurea's twitter unbeknownst to him.
professor fennel: AHHH I LOVE HER!! i ship her and juniper tbh... i like the idea that they met and started dating in college and are still in a relationship of some kind! interesting because i hc'd juniper as a evolutionary biologist and fennel would probably be somewhere in the realm of neuroscience, which are pretty different (unless we're talking about the evolution of the brain and stuff). so how would they have met.... something i'd like to think about further. to parallel with gamesync, i feel like she's probably really good at coding things like python(? i'm not a code person. yet,,). i know nothing about neuroscience but maybe she researches ways we can better understand dreams by emulating things going on in the brain during sleep into computer ai's or algorithms. like a computer model? i'm riffing at this point. also maybe she also knows r and juniper would make excuses for them to hang out by begging her to teach her how to use r. neuroscience/coding tumblr pls do not kill me
professor ivy: i didn't know who this was and had to look it up and ohhh it's the sexy professor people were talking about from the anime. MILF ok wow the fact that the anime went as far to make up a paper she's written, "Pokémon adaptive variation as a function of regional distribution," is REALLY impressive. that actually makes me really excited! (i hope i'm about to explain this right) adaptive radiation is the process in which a group of organisms speciates at a relatively fast rate because of some incredibly useful innovation, like invading a new underrepresented niche (environmental role/place) or developing a useful novel (new) trait. an example would be when the dinosaurs went extinct, the ancestors of mammals were able to diversify from being small insect-eaters to big carnivores, herbivores, generalists, and more (the dinosaurs that were the ancestors of birds were also really small and not diverse so they were able to radiate into what is now known as birds too). describing adaptive radiations as a function of regional distributions is making me think it's the former, that as pokemon disperse, they invade new types of habitats that are relatively uninhabited and eventually evolve adaptations to those environments, eventually resulting in new species. or maybe they were able to outcompete natives. just "pokemon" is really broad though so maybe the paper is a kind of literature review that summarizes papers talking about specific instances of dispersal and speciation, makes suggestions for future research, and discusses theories on adaptive radiation in terms of dispersal patterns and geological events. honestly i'm really astounded the anime was able to come up with that title, i declare philena ivy the biggest brained out of all the professors. definitely another big-name evolutionary biologist. OH WAIT SHE LIVES ON AN ISLAND?? she DEFINITELY studies island endemics (organisms found only in one area in the world). i know lots of people who study island endemics- my mammal diversity professor, my lab pi who studies snails and all her grad students who do too, and anyone who studies anole evolution.
#sorry if there's any factual errors or oversimplifications i wrote this really fast at 1 am#pokemon#cedric juniper#professor fennel#professor ivy#philena ivy#ask#lab notebook
3 notes
·
View notes
Text
PhD as a job
I recently reread this passage in David Graeber's _Bullshit Jobs_ (by the way, I highly recommend reading the book):
In the spring of 2013, I unwittingly set off a very minor international sensation.
It all began when I was asked to write an essay for a new radical magazine called Strike! The editor asked if I had anything provocative that no one else would be likely to publish. I usually have one or two essay ideas like that stewing around, so I drafted one up and presented him with a brief piece entitled “On the Phenomenon of Bullshit Jobs.”
The essay was based on a hunch. Everyone is familiar with those sort of jobs that don’t seem, to the outsider, to really do much of anything: HR consultants, communications coordinators, PR researchers, financial strategists, corporate lawyers, or the sort of people (very familiar in academic contexts) who spend their time staffing committees that discuss the problem of unnecessary committees. The list was seemingly endless. What, I wondered, if these jobs really are useless, and those who hold them are aware of it? Certainly you meet people now and then who seem to feel their jobs are pointless and unnecessary. Could there be anything more demoralizing than having to wake up in the morning five out of seven days of one’s adult life to perform a task that one secretly believed did not need to be performed—that was simply a waste of time or resources, or that even made the world worse? Would this not be a terrible psychic wound running across our society? Yet if so, it was one that no one ever seemed to talk about. There were plenty of surveys over whether people were happy at work. There were none, as far as I knew, about whether or not they felt their jobs had any good reason to exist.
This possibility that our society is riddled with useless jobs that no one wants to talk about did not seem inherently implausible. The subject of work is riddled with taboos. Even the fact that most people don’t like their jobs and would relish an excuse not to go to work is considered something that can’t really be admitted on TV—certainly not on the TV news, even if it might occasionally be alluded to in documentaries and stand-up comedy. I had experienced these taboos myself: I had once acted as the media liaison for an activist group that, rumor had it, was planning a civil disobedience campaign to shut down the Washington, DC, transport system as part of a protest against a global economic summit. In the days leading up to it, you could hardly go anywhere looking like an anarchist without some cheerful civil servant walking up to you and asking whether it was really true he or she wouldn’t have to go to work on Monday. Yet at the same time, TV crews managed dutifully to interview city employees—and I wouldn’t be surprised if some of them were the same city employees—commenting on how terribly tragic it would be if they wouldn’t be able to get to work, since they knew that’s what it would take to get them on TV. No one seems to feel free to say what they really feel about such matters—at least in public.
It was plausible, but I didn’t really know. In a way, I wrote the piece as a kind of experiment. I was interested to see what sort of response it would elicit...
[Graeber reprints his poignant essay]...
If ever an essay’s hypothesis was confirmed by its reception, this was it. “On the Phenomenon of Bullshit Jobs” produced an explosion.
It occurred to me that, just as "bullshit jobs" was taboo to discuss but of wide resonance, the same might apply to **the steps one should take to pick, perform and/or leave jobs if one has internalized that one's job or career is indeed bullshit.**
At least it seems that way in my university's PhD programs. I suspect many of my peers don't like their PhD jobs, and distinctly, many have jobs they "secretly believe [do] not need to be performed." I suspect many of those peers are actively interested in how to deal with these situations: should I leave PhD? If not and if I haven't yet chosen a boss, how should I choose? If I'm already stuck with a boss, should I switch? Should I try shirking and see what happens? Should I just keep working as I am and stop worrying about the utility of my work, which could very possibly make me happy? Will bringing in my own funding give me more negotiating power? I'll call these questions "bullshit job considerations." But it's taboo—I haven't found students talking about these questions, and almost always I'm the first one to bring up that I've even _considered_ these questions (which evokes one of a few reactions: giggles, "don't say that!" or rarely, "yeah maybe I should think about that too") before meaningful discussion ensues.
This is not to say that there aren't many students who enjoy their PhDs: it seems highly variable. On the question of the usefulness of their jobs, I suspect many students would say that they feel they're contributing to knowledge, teaching others and helping advance health and medicine.
But for those students who don't feel so positive, I think it could be very helpful to open up spaces in which to discuss what to do in reaction to those feelings, to make this area of discussion and thinking non-taboo. For example, many students entering PhD are confronted with the decision of which boss and lab to work for in their thesis research. Each PhD program, older PhD student and postdoc will recommend criteria with which to make this decision. Commonly, I've heard "Make sure your PI [principal investigator, aka boss] and you get along," or "Look for a PI has your interests at heart." I agree that these are important criteria, depending on the definition of "your interests." I think these criteria would be greatly supplemented by "Look for a PI who gives you ample free time," "Look for a PI and lab that don't believe in 'work as an end and meaning in itself' (Graeber), e.g. a PI who won't be happy or sad with you depending on the number of analyses or experiments you run," or "Look for a PI who won't pressure you into working on a project whose usefulness you don't understand or believe, even if that means long spells of no visible production." I myself am unsure whether I can realistically find a PI matching these criteria, given that my university research environment feels very productivity-focused, but I think these criteria are at least worth acknowledging as things to aspire to.
Talking about leaving the PhD is even more taboo, although surprisingly, I've heard more talk about that than about tactics for increasing free time, either by shirking (which seems to be more commonly discussed and practiced in other industries) or by winning negotiating power with your boss by "providing value" to them. The latter is a major subject in multiple books, including Tim Ferriss's _4 Hour Workweek_, which enjoys popularity outside my university's academic bubble. (This again deepens my stereotype that we have a longer way to go on these issues in academia.)
I sometimes feel, inspired by Nassim Taleb, that a very good work setup for me would be either complete freedom (i.e. retired and pursuing my own projects) or sinecure-cum-freedom (i.e. having a job that disappears after I leave the office at 12pm each day, and complete freedom afterwards)[1][2]. Another good setup would be in an organization I really believed in (a combination of many factors) and could contribute to. At this point, these feel mostly compatible with my PhD, pending finding a compatible boss, but I haven't seriously imagined if life not in the PhD would be better for pursuing these.
Footnotes:
I've experienced complete freedom and I think that some balancing force of commitment, i.e. the sinecure-cum-freedom, might actually feel better. It's hard for me to pinpoint exactly why—perhaps some routinized social interaction or feelings of immediate usefulness to your co-workers.
What counts as a "very good work setup" probably depends a lot on the person, but at the same time, I do think many people look for similar things. I think control and freedom over one's time and one's self is a common value, and I think the work setups I mention above are trying to honor exactly that value.
1 note
·
View note
Text
CHARACTER INTERVIEW.

NAME: marigold summer thompson NICKNAME: mari ; gold ; goldie locks ; mother nature ; earth child ; witch’s kid AGE: 17 ( s1 ), 18 ( s2 ) SPECIES: human with powers
personal.
MORALITY: lawful / neutral / chaotic / good / neutral / evil RELIGIOUS BELIEF: raised in a lab, later rejoins hippie parents; father is not religious but mother is wiccan (pagan and a witch) SINS: greed / gluttony / sloth / lust / pride / envy / wrath VIRTUES: chastity / charity / diligence / humility / kindness / patience / justice PRIMARY GOALS IN LIFE: to live. she has spent the majority of her childhood in a lab, being tested on, then was thrown back to her parents when they had no use for her. she just wants to live her life. due to her powers, she’ll stay in an environment-based field. like her parents, she’ll be a “tree hugger” and be an accomplished horticulturist. LANGUAGES KNOWN: english, “nature language” SECRETS: hawkins lab test subject. she was kept in the lab for her whole childhood which made her not experience pop culture for those years. she tries to get away with not understanding references, but sometimes she is just utterly confused. also, despite her parents being hippies and her kind demeanor, if someone comes at her with insults about her or her family, she will bite back twice as hard.
BUILD: scrawny / bony / slender / fit / athletic / curvy / herculean / pudgy / average HEIGHT: 5′10′‘ WEIGHT: 121 lbs SCARS/BIRTHMARKS: various scars on her body from testing and injuries from her time at hawkins, 001 on her left forearm from hawkins lab ABILITIES/POWERS: elemental manipulation. the ability to manipulate and communicate with the four elements and nature. RESTRICTIONS: horrible people, people who make fun of her family
favourites.
FOOD: chicken pot pies….don’t tell her parents DRINK: orange soda PIZZA TOPPING: basil leaves and tomatoes COLOUR: sunset MUSIC GENRE: she likes new-age because of her parents, but she also likes a bit of rock and folk music, stevie nicks is her idol. BOOK GENRE: ones that tell the complexity and intricacies of life MOVIE GENRE: comedies, thought-provoking SEASON: summer, it’s when all of the elements are the happiest CURSE WORD: shit, fuck SCENT(S): pine, lavender, jasmine
fun stuff.
BOTTOM OR TOP: bottom most likely, unless she is in a situation with someone where she feels an overwhelming feeling of lust…she’ll pin them SINGS IN THE SHOWER: every single time! LIKES BAD PUNS: she does. very much.
tagged by: i took it upon myself to do it bc i saw it on my dash
tagging: anyone!!!
2 notes
·
View notes
Text
First Take Review: RME ADI-2 DAC FS

I learned about the RME DAC when stumbling upon The Master Switch's "Best DAC" list. I don't remember what led me there, but from that list it sounded like it stacked up well against some other popular audiophile DACs from the likes of Mytek, Schiit, Benchmark and Chord. After a bit of online research I was intrigued by the internet buzz around it, and its pro-audio heritage, ostensible quality (made in Germany!) and unique feature set. And at USD $1,149, it hits a price point that many audiophiles can stomach. This is not another $299 made-in-China special off Amazon - not that those can't sound good or be amazing values, but the RME is something entirely different.
RME is a German manufacturer of highly-regarded pro audio gear that decided to make a version of their studio-grade ADI-2 DAC for the audiophile market. This meant equipping it with standard consumer digital inputs, adding dual headphone amps for both big high-impedance cans and low-impedance IEMs, making the menu system simpler (it's still rather arcane in that German first-gen-BMW-iDrive kind of way), making the informative but busy display automatically dimmable and throwing in a remote control. I'll spare the technical details for the full review, but highlights include a very precise reclocking circuit called SteadyClock FS, AKM4493 DACs and three stages of internal power regulation (switching, linear and ultra low-noise) that purportedly make it immune to the quality of the external power supply, the included one being be a run-of-the-mill 12V 2A brick. Other than the dinky supply, the ADI-2 feels extremely sturdy and is clearly designed to withstand the rigors of studio and field use, something you certainly can’t say of the vast majority of audiophile gear. The rotary controls are firm and tactile, and everything feels designed for the highest level of utility and durability with zero fluff.

Decidedly consumer-friendly connections
Despite being simplified from the original Pro model, this thing is absolutely packed with processing features. Adjustable digital filters, parametric EQ, tone controls, loudness contour, crossfeed for headphones - you name it, it's probably got it. There’s no network connection though, so for streaming you’ll need to provide your own USB or S/PDIF device. It has a full digital volume control, so if you're ok with some digital manipulation (albeit done at very high precision), you can hook it up directly to an amp or active speakers and have volume control via the full-function remote. The analog output can be set to 4 different gain settings via the menu system, and I found the +7dbU setting to come closest to my PS Audio DirectStream via XLR output. I kept the digital volume fixed at 0dB.
Setup ⚙️
My initial test was using a Raspberry Pi 3B with Roipeee software as a Roon endpoint, connected to the RME via DH Labs Mirage USB cable ($240). I listened on my main rig, currently consisting of a PS Audio DirectStream DAC, Pass Labs XP10 preamp, Pass Labs XA30.5 or Valvet A4 Mk.II amplifiers, Audiovector SR 6 Avantgarde Arreté speakers and cables and power conditioning from Furutech and Audience. (Yes, I know, a $50k system isn’t the most logical pairing for a ~$1k DAC, so I’ll also be reporting later on results in my more proportionately-priced side system.)
I am not a headphone guy at all, much preferring to listen to everything on my big rig, so I'm not qualified to critically evaluate the headphone amps and will defer to the other reviews on the 'net. That said, I did some light testing with my Ultimate Ears TripleFi IEMs and Massdrop HD6xx, which is a Sennheiser HD650 reproduction (I'll just refer to it as a HD650 for simplicity). For light comparison, I have my Topping NX4 DSD portable USB DAC + headphone amp which cost around $150 when I bought it and is insanely good for the money. I’ve also owned an Ayre Codex DAC + headphone amp ($1795) and remember its sound pretty well, but no longer have it on hand.
An important note: all my speaker listening was performed using the RME's balanced XLR outputs, as both my main and side systems are fully balanced (Pass Labs in the main rig, Ayre on the side). This is of course the preferred connection method in pro audio environments as well, and the RME is designed with fully balanced operation in mind. I have not listened to the RCA outputs yet, but my gut tells me balanced is going to be the way to go (and this account seems to back that up). So keep that in mind - I'll explore the sound of the unbalanced outs in a later review.

A “Dark Mode” setting auto-dims the informative but garish display
The Sound 🔉🎶
First impressions: clean, clean, clean. Excellent resolution, smooth and neutral tonality. Very clear without being bright or clinical. Good crisp dynamics. Everything sounds in its proper place without obvious artifice. Not quite as fluid and relaxing as the PS Audio DirectStream ($5,999), but quite enjoyable on a high resolution system.
Second impressions: I moved the RME to my office area for headphone listening, connected it to my MacBook pro via the same USB cable. Keeping in mind my prior disclaimer that I'm not a headphone guy, I found the RME to again sound very clear and clean on both the HD650's and Ultimate Ears TripleFi's, but maybe lacking a little grunt. While the DAC quality was clearly a step above the Topping NX4 DSD in resolution and transparency, I found the RME a little subdued, both via the IEM output with the TripleFi IEMs and the high power output with the HD650. The latter tends towards mellowness and needs an amp with some moxie to bring it alive; the RME didn't strike me as that, sounding as if it needed a beefier power supply or perhaps a higher-voltage output stage that could slew a little harder. And I found the Topping to give the Ultimate Ears a bit more drive to the sound, though that might also be the Topping erring on the punchy side of neutral. Again, I would hold judgement here, as I only did brief tests and this could also be a function of my digital front-end.
Third impressions: Fast forward a couple weeks, and I moved the RME back to the main rig, but this time with a new Raspberry Pi 4 set up as the Roon endpoint. I have not optimized my streaming setup in the least, but my understanding is the added power and USB bandwidth of the rPi 4 should translate to better performance. And it certainly sounded that way - the very slight grain from my first go with the rPi 3B was diminished, and the sound had greater scale and ease. While I still consistently preferred the PS Audio, it was getting harder tell them apart in quick A/B tests. There's a subliminally less "digital" quality the the PS Audio - it is still the most natural DAC I have had in my system - and everything seems to sing just a bit more freely through it. The RME comes fairly close but sounds a bit flatter and less tactile. Despite the S/N specs being significantly better on the RME, I didn't find it to be absolutely quieter - some aspects felt a little cleaner, but the PS Audio has a better sense of blackness between the notes. The RME is a bit more up-front with its soundstage vs. the PS Audio which put things a hair further back, but with a more natural sense of space. Still, I find the RME less in-your-face and more relaxed than many of the Sabre-based DACs I've heard.
In a nutshell 🥜

So, how do I think the $1,149 RME stacks up against "serious" audiophile DACs like the PS Audio DirectStream at 4x the price? Short answer: the DirectStream isn't going anywhere. But the RME comes much closer to the $6k PS Audio than I was expecting, particularly in terms of tonality, dynamics and resolution. The main differences I hear are the PS Audio has a bit more weight (both dynamically and tonally), a better sense of space and tactility and a less "electronic" overall sound. The RME makes symphony orchestras sound a tad smaller, with less of a difference in scale between a solo oboe and a string section... qualities that are pretty subtle and might not matter to some, but are highly prized by those of us seeking that visceral connection to our reproduced music. This is generally what you have to pay the big bucks for, and what puts DACs like the PS Audio, the Chord DAVE, the MSB’s and the Mergings in a different class from the more affordable offerings. But the RME acquits itself extremely well and is a nice upgrade in naturalness from other very good DACs that I’ve heard at the price point, e.g. from Cambridge Audio. And from aural memory, I feel the RME is a more accurate and satisfying DAC than the Ayre Codex, which never came as close to matching my DirectStream.
I'll be delving more into this fascinating little product in the coming weeks, but you can add this to the internet buzz: the RME ADI-2 FS DAC is an excellent sounding piece worthy of audiophile credibility. And I haven't even begun to tap into all its unique capabilities that no competitor I'm aware of can offer. Before you spend more on a DAC, you may want to give this little guy a listen.
0 notes
Text
Student Spotlight: Megan Bazin, Integrated Pest Management Intern
My name is Megan Bazin and I just graduated from Temple University with a BS in Horticulture. I'm from South Jersey and am the year-long Integrated Pest Management intern here at Longwood.
What is your favorite plant?
My favorite plant is the white pine, because I used to climb them a lot as a kid. I also really love their form when they mature. Being from the pine barrens, the smell of pine trees is very nostalgic for me.
What is your favorite Garden? What is your favorite part of Longwood?
My favorite garden is the arboretum at the Temple University Ambler Campus. I worked there for 3 years while I completed my degree, and it has a lot of sentimental value for me. My favorite part of Longwood is either Pierce's Woods or the Hillside garden. I like native perennials and natural-feeling environments.
What is the best part of being a student?
Being able to ask as many questions as I want. At many jobs, you're expected to just do your work as quickly as possible and not ask questions. When you're a student, you have the opportunity to understand why you're doing what you're doing; what the bigger picture is. Understanding the bigger picture means you can come up with your own innovative ideas and creative solutions to problems.
What is your background in horticulture (or whatever field you are in)?
I have done various internships and volunteer programs in the fields of horticulture, urban agriculture, and organic farming in addition to my four years at Temple University Ambler. At Ambler, I was the Native Plant Intern, where I took care of our Native Formal Garden and helped catalog plant records. I also cared for and forced plants for Temple's Philadelphia Flower Show exhibit for three years in a row.
Why did you want to come to Longwood and what do you think helped you get the position?
I wanted to come to Longwood because I wanted a specialization in addition to my general education in horticulture. Longwood's year-long internship position was a perfect opportunity for me. I chose the IPM specialization because it's applicable to a variety of areas within the field, but also because plant pathology and entomology were my favorite classes in college. I think I got the position because I interviewed well, and had good recommendations. Longwood and Temple work together often, so I think my being a senior horticulture student there was a bonus.
What do you do at work?
My favorite thing to do at work is when I get to research a problem about a plant and find out a solution to the problem. I like being a scientist and feeling like I am really contributing to our stewardship of plants. Sometimes this involves using a map and accession number to locate a plant, and I feel like I'm treasure hunting, which is especially fun.
What are your future plans or what is your intended career path?
I would be happy with any job working with plants and the outdoors. I really enjoy IPM, so I am hoping to utilize this specialization.

Temple's 'Next Stop' interview in my beloved arboretum.

Diagnosing plant diseases in the Longwood IPM lab.

Water Lilies field trip into the ponds!

Installing a garden with fellow Horticulture Honor Society members (Pi Alpha Xi).

Getting very excited about the china during a trip to the Hagley Museum.
#student#student spotlight#megan#IPM#intern#internship#horticulture#Longwood Gardens#Temple University Ambler
1 note
·
View note
Text
Aimless
I’ve made it. I’ve done it. I’ve reached my goals. Did I really want to reach team?
I am struck by the futility of life and action. What’s the point? Why do I try? What am I trying to do? Perhaps this is a mini-mid-life crisis brought on by COVID, or perhaps it’s something that has been in the back of my mind once I saw what the result of my goals actually looks like.
Do I want to work on optimization code to make robots marginally better at processing something when it will likely be overtaken by advancements in processors in the near future? Do I want to work on optimizing some AI system whose performance will easily be overtaken by the latest paper? Do I want to spend all my time building some advanced architecture which will most likely be discarded like all the rest in the future? What’s the point?
For the longest time my goal was to learn all the skills I thought I needed to do robotics. I have learned a lot of them--programming, AI, 3D modelling, 3D printing, electronics, some machining, some control systems...and yet, something is still missing. I work at NASA and I am more unhappy about the work I am doing than I have been in a while. Maybe this is just the nature of research work--we spend a whole lot of time trying to do something only for it ultimately to be something thrown away. Only the 0.1% of research that makes breakthroughs is ultimately useful to the world.
Why am I unhappy? I don’t enjoy the work I am doing. I don’t like my work environment. What’s wrong with my work? ... Well my whole reason for working with robots was to see them actuate in real life. I don’t want to be pushing algorithms behind the scenes and never see my work doing something. I’m building up a toolset that will likely only ever be used by me and perhaps one or two others. Worse, I’m making marginal improvements to a system whose performance does not seem to be getting any better than my initial results despite all the time and effort I have put into it.The fact that I don’t get to work in person with anyone or with any actual robot is soul crushing. This ties in with the work environment--working alone sucks. Telework is nice once in a while, but I think actually going in and working alongside people is so much more motivating, so much more satisfying. It’s easier to balance my time and I gain a lot more knowledge just by being in the vicinity of smart people and hearing about their work.
I have difficulty feeling motivated working on my own stuff of my own volition unless there’s some particular “cool” aspect that I really enjoy--even then, there’s usually a lull where the sheen fades and I just have to keep trudging on in order to finish the job. I guess I enjoy that part right after starting where I start to figure things out and I get a result, but not the part where I polish it into something reusable and desirable for others. The prototyping stage is the most fun part of any project.
Unfortunately the reality is that my work is going to be like this for a while longer. Maybe even a lot longer based on the way this quarantine is shaping up. So how can I make it work? How can I adapt to these shitty circumstances while still being happy, healthy, and productive?
General wisdom says that a mindset of gratitude is the key. Perhaps it’s true that I haven’t been grateful enough. I am at one of the top universities in the world working with some of the greatest minds. I have plenty to eat, stable finances, a patient family, and friends I can call if I really feel the need. I’m not worried about survival at this point, only about fulfillment--many in the world cannot say this. There are probably 1000′s if not millions of people who would love to be in my position.
I am grateful for all these blessings, but I also feel almost soul-crushingly responsible because of them. Like if I don’t do something great, I am betraying all those who have raised me up to this point. If I waste my time I am insulting those that have sacrificed for my sake. It feels like too much sometimes, and I just want to give up, to go on an extended vacation, and just be free for a while. The thing is, I feel obligated to work 40 hrs a week, and if I don’t do that, I’m worried that I am robbing the people paying me. One could argue that my time is more valuable, that I’m being underpaid, or that I’m more efficient than the average, but this doesn’t change the fact that I made a contractual agreement to put in the time.
So what do I do? How do I demonstrate my gratitude? How do I stay productive with work while also finding fulfillment? Is there room for me to work on my own projects that I enjoy more? Do I even enjoy my own projects? What do I want to do? The question of goals comes to the forefront of my mind again. Perhaps an overarching goal and provide an underlying drive to everything I do.
Do I want to go into research? My current view of research in my field is that I would spend a whole lot of time on my own working on something of debatable value and unproven possibility anchored to the underlying goals of my selected PI. The other possibility is that if I had a strong enough idea and open enough PI, I could probably shape my work to something I believe would work, find a group of people that I believe are capable, and then use that as a launching pad for building a company, maybe not even completing my PhD.
What am I good at? What do I like? I am good at building and understanding large systems/frameworks/architectures. I have a variety of shallow skills, but my only real specialization is software engineering. My math is OK, physics is decent, electronics is decent, 3D modeling, fabrication, and mechanical mindset is mediocre, AI is okay, though I doubt I would make my own network or algorithm there, and my leadership skills are alright. I am also decent at teaching (or at least patient enough to learn to do it well). I feel I would thrive in an environment where I get other people started with something new and they take it to its ultimate fruition. Either that, or leading a small team after getting a prototype out. System architect would be okay, though I would like to work with hardware too. Really what I want is to replicate the environment I usually see in maker spaces and robotics clubs when people are working on the same project. It’s open, free, laidback, and fun. Everyone gets to share in the successes and failures and just screw around a little bit. I suppose the question is how I can replicate that kind of environment in the real world while still making enough money to support everyone involved? A startup seems like the most natural conclusion. Either one I start or one I find that has the culture I like. I think the main thing is I don’t really want to work on projects alone or with people who have significantly less investment.’
Aside: The VR project I did was really cool in terms of team structure. It sort of fell apart due to COVID, but if we had had the space and in-person time I would have liked, it could have been much better; 3/4 of the team was on top of things too, which was excellent (The other 1/4 was a waste of my time, but what can you do).
Okay, so the goal is to find a way to replicate the environment of robotics club somehow while making something valuable. This means that wherever I go, I need to work somewhere where collaboration is core, and not just an afterthought where each person works in a silo and then meets together after 100 hours. The Interaction Lab was kind of this, but they had the issue of unequal investment. Investment should be 2:1 at worst, and ideally it would be 1:1 for everyone. A 4:1 ratio or greater is just unproductive unless the 1 is a consultant/expert/advisor rather than a regular team member.
The question then becomes: what do I need to do or learn in order to make this work environment a reality? I suppose one option is to just go around and meet a bunch of different teams and see how they work, then attempt to join them. The other approach is I create my own team using startup money or research funds. The former option requires an idea and startup mentor; the latter requires a hypothesis and a PI. Both require me to find people that would join me. The challenge for me is likely that for me the work environment is central, but for many others it is the idea that is central. While the idea is very important, I’d say the work environment is much more important. A good team can produce value very easily with many ideas, while a poor team with a fantastic idea will struggle to produce value. That said, usually it’s the idea/vision which motivates good people to join a team. I need to make a point to keep up an idea journal again. The best way to enact my vision for a working environment is to start it myself, but to do that I need a vision people will get behind along with their trust.
So what are the other goals, and how will I adapt to COVID in the short term? Personal health is one that comes to the forefront--I’ve been living unhealthfully for a while now; my work should never be more important than my health (with some rare exceptions perhaps). Secondary to that is relationships. I realize now that sitting around until I finish XYZ before looking for a significant other is pointless. Yes, I will “lose” a fair bit of time courting someone, but ultimately it will be worth it once I find the right person, and there’s no real reason for me to hesitate. I will never reach the absolute ideal of physical performance or personal life habits, and must instead bank on love developing between flawed individuals.
COVID adaptation: For work, separating it into two working sessions per day is probably the key. Requiring any more than that does not feel sustainable or valuable right now. Focusing on hitting 8 hours every day feels like a slog when I’m by myself. I know I can accomplish enough work in less time if I’m focused. I’ll probably be more effective this way as well since i won’t burn out. After or between those working sessions, I should work on personal passion projects which I feel will make me better at something in some way. These will make me feel some sense of progress every day even if work does not provide it for whatever reason. For exercise, I just need to do it. The time of the day it occurs in doesn’t really matter, I just need to get outside and be at least somewhat active.
Aim acquired. Thanks God.
1 note
·
View note
Text
Research summary - Predicting phenotypes of individuals based on missense variants and prior knowledge of gene function
I have been meaning to write blog posts summarising different aspects of the work from our group over the past 6 years, putting it into context with other works and describing also some future perspectives. I have just been at the CSHL Network Biology meeting with some interesting talks that prompted me to put some thoughts to words regarding the issue of mapping genotypes to phenotypes, making use of prior cell biology knowledge. Skip to the last section if you just want a more general take and perspective on the problem.
Most of the work of our group over the past 6 years has been related to the study of kinase signalling. One smaller thread of research has been devoted to the relation between genotypes and phenotypes of individuals of the same species. My interest in this comes from the genetic and chemical genetic work in S. cerevisiae that I contributed while a postdoc (in Nevan Krogan’s lab). My introduction to genetics was from studies of gene deletion phenotypes in a single strain (i.e. individual) of a model organism. Going back to the works of Charlie Boone and Brenda Andrews, this research always emphasised that, despite rare, non-additive genetic and environment-gene interactions are numerous and constrained in predictable ways by cell biology. To me, this view of genetics still stands in contrast to genome-wide association studies (GWAS) that emphasise a simpler association model between genomic regions and phenotypes. In the GWAS world-view, genetic interactions are ignored and knowledge of cell biology is most often not considered as prior knowledge for associations (I know I am am exaggerating here).
Predicting phenotypes of individuals from coding variants and gene deletion phenotypes
Over 7 years ago, some studies of strains (i.e. individuals) of S. cerevisiae made available genome and phenotypic traits. Given all that we knew about the genetics and cell biology of S. cerevisiae I thought it would not be crazy to take the genome sequences, predict the impact of the variants on proteins of these strains and then use the protein function information to predict fitness traits. I was brilliantly scooped on these ideas by Rob Jelier (Jelier et al. Nat Genetics 2011) while he was in Ben Lehner’s lab (see previous blog post). Nevertheless, I though this was an interesting direction to explore and when Marco Galardini (group profile, webpage) joined our group as a postdoc he brought his own interests in microbial genotype-to-phenotype associations and which led to a fantastic collaboration with the Typas lab in Heidelberg pursuing this research line.
Marco set out to scale up the initial results from Ben’s lab with an application to E. coli. This entailed finding a large collection of strains from diverse sources, by sending emails to the community begging them to send us their collections. We compiled publicly available genome sequences, sequence some more and performed large scale growth profiling of these strains in different conditions. From the genome sequences, Marco calculated the impact of variants, relative to the reference genome and used variant effect predictors to identify likely deleterious variants. Genomes, phenotypes and variant effect predictions are available online for reuse. For the lab reference strain of E. coli, we had also quantitative data of the growth defects caused by deleting each gene in a large panel of conditions. We then tested the hypothesis that the poor growth of a strain of E. coli (in a given condition) could be predicted from deleterious variants in genes known to be important in that same condition (Galardini et al. eLife 2017). While our growth predictions were significantly related to experimental observations the predictive power was very weak. We discuss the potential reasons in the paper but the most obvious would be errors in the variant effect predictions and differences in the impact of gene deletion phenotypes in different genomic contexts (see below).
Around the same time Omar Wagih (group profile, twitter), a former PhD student, started the construction of a collection of variant effect predictors, expanding on the work that Marco was doing to try to generalise to multiple mechanisms of variant effects and to add predictors for S. cerevisiae and H. sapiens. The result of this effort was the www.mutfunc.com resource (Wagih et al. MSB 2018). Given a set of variants for a genome in one of the 3 species mutfunc will try to say which variants may have an impact on protein stability, protein interactions, conserved regions, PTMs, linear motifs and TF binding sites. There is a lot of work that went into getting all the methods together and a lot of computational time spent on pre-computing the potential consequence of every possible variant. We illustrate in the mutfunc paper some examples of how it can be used.
Modes of failure – variant effect predictions and genetic background dependencies
One of the potential reasons why the growth phenotypes of individual stains may be hard to predict based on loss of function mutations could be that the variant effect predictors are simply not good enough. We have looked at recent data on deep mutational scanning experiments and we know there is a lot of room for improvement. For example, the predictors (e.g. FoldX, SIFT) can get the trends for single variants but really fail for more than one missense variant. We will try to work on this and the increase in mutational scanning experiments will provide a growing set of examples on which to derive better computational methods.
A second potential reason why loss of function of genes may not cause predictable growth defects would be that the gene deletion phenotypes depends on the rest of the genetic background. Even if we were capable of predicting perfectly when a missense variant causes loss of function we can’t really assume that the gene deletion phenotypes will be independent of the other variants in the genome. To test this we have recently measured gene deletion phenotypes in 4 different genetic backgrounds of S. cerevisiae. We observed 16% to 42% deletion phenotypes changing between pairs of strains and described the overall findings in this preprint that is currently under review. This is consistent with other works, including RNAi studies in C. elegans where 20% of 1,400 genes tested had different phenotypes across two backgrounds. Understanding and taking into account these genetic background dependencies is not going to be trivial.
Perspectives and different directions on genotype-to-phenotype mapping
Where do we go from here ? How do make progress in mapping how genotype variants impact on phenotypes ? Of course, one research path that is being actively worked on is the idea that one can perform association studies between genotypes and phenotypes via “intermediate” traits such as gene expression and all other sorts of large scale measurements. The hope is that by jointly analysing such associations there can be a gain in power and mechanistic understanding. Going back to the Network Biology meeting this line of research was represented with a talk by Daifeng Wang describing the PsychENCODE Consortium with data for the adult brain across 1866 individuals with measurements across multiple different omics (Wang et al. Science 2018). My concern with this line of research is that it still focuses on fairly frequent variants and continues not to make full use of prior knowledge of biology. If combinations of rare or individual variants contribute significantly to the variance of phenotypes such association approaches will be inherently limited.
A few talks at the meeting included deep mutational scanning experiments where the focus is mapping (exhaustively) genotype-to-phenotype on much simpler systems, sometimes only a single protein. This included work from Fritz Roth and Ben Lehner labs. For example, Guillaume Diss (now a PI at FMI), described his work in Ben’s lab where they studied the impact of >120,000 pairs of mutations on an protein interaction (Diss & Lehner eLife 2018). Ben’s lab has several other examples where they have look in high detail and these fitness maps for specific functions (e.g. splicing code, tRNA function). From these, one can imagine slowly increasing the system complexity including for example pathway models. This is illustrated in a study of natural variants of the GAL3 gene in yeast (Richard et al. MSB 2018). This path forward is slower than QTL everything but the hope would be that some models will start to generalise well enough to apply them computationally at a larger scale.
Yet another take on this problem was represented by Trey Ideker at the meeting. He covered a lot of ground on his keynote but he showed how we can take the current large scale (unbiased) protein-protein functional association networks to create a hierarchical view of the cellular functions, or a cellular ontology (Dutkowski et al. Nat Biotech 2013 , www.nexontology.org). Then this hierarchical ontology can be used to learn how perturbations of gene functions combine in unexpected ways and at different levels of the hierarchy (Ma et al. Nat Methods 2018). The notion being that higher levels in the hierarchy could represent the true cellular cause of a phenotype. In other words, DNA damage repair deficiency could be underlying cause of a given disease and there are multiple ways by which such deficiency can be caused by mutations. Instead of performing linear associations between DNA variants and the disease, the variants can be interpreted at the level of this hierarchical view of gene function to predict the DNA damage repair deficiency and then associate that deficiency with the phenotype. The advantages of this line of research would be to be able to make use of prior cell biology knowledge and in a framework that explicitly considers genetic interactions and can interpret rare variants.
I think these represent different directions to address the same problem. Although they are all viable, as usual, I don't think they are equally funded and explored.
— Evolution of Cellular Networks
#Evolution of Cellular Networks#Research summary - Predicting phenotypes of individuals based on mis
0 notes
Note
how did you go about finding a lab, PI and a grad school? I’m kind of confused by the process. I’ve been told it doesn’t matter what the lab/degree is as long as it corresponds to what you want to do research in.. other times I’ve been told to select a mentor not a project? And I honestly am so confused I don’t even know how to properly begin and my undergrad advisor was of little help.
Hello!
I definitely understand. The process IS pretty overwhelming. Hopefully I can help.
A disclaimer first: this advice may only be applicable for life science PhD programs in the United States (as that’s what I’m most familiar with). If you’re applying for another program or degree, in another country, some of my advice may not apply because they may have different processes.
My process went like this: apply for and interview at grad school program -> get accepted to that university and program -> rotate through 3 different labs -> choose the lab/PI that worked best for me. (Some countries the process is reversed--you apply to the lab, then after you get accepted to the lab, you gain acceptance into the university).
My advice on narrowing down what grad schools to apply to
Advice on choosing a lab
Potential questions to ask your potential PI to see if the lab is right for you
Guide on PhD lab rotations
As for your specific questions/statements:
I’ve been told it doesn’t matter what the lab/degree is as long as it corresponds to what you want to do research in
The answer is kind of multi-faceted and long-winded and very “it depends”.
Your lab/degree/program dictates your research, and your research project may shape the networks you interact with in the field, and your networks may influence what post-grad opportunities are offered to you (eg. a post-doc or industry position). This is especially true for those wanting to go into academia, where everything you do as a whole matters more.
However, you can also create networks that open up other doors outside of your immediate field. For example, an alumnus from my program got a job as an editor at the journal Cell after graduating--not because of her research per se, obviously, but because she was able to publish a ton of papers (on her research) and was really familiar with the whole process of scientific writing/editing/publishing/etc, and thus gained a lot of network connections in the publishing field.
Another case: my lab mate is currently doing a post-doc in a pretty different subject than what his grad project (and degree) is in. He has a PhD in Nutritional Sciences, his PhD project was on drug metabolism, but his post-doc now is on cancer biomarker detection.
That being said, in some cases, your degree (ie. PhD) is sometimes more important than your dissertation project. But in other cases, your degree is also the first thing employers see and thus make their first impressions on. So if I, a Cancer Biologist, tried to apply for a job at a Physics-heavy company, they may pass over me for an actual Physicist. My Nutritional Scientist buddy had kind of a hard time when applying to cancer research companies, despite his stellar knowledge on cancer biology (coming from a cancer biology lab), because all HR saw were the two words by his PhD.
So ya, long answer is: yes, no, sometimes, it depends.
other times I’ve been told to select a mentor not a project?
This part is absolutely true. Your project does not matter one bit if your mentor creates a toxic environment for you that makes you want to quit grad school. I think I say this in one of those links up there but your grad school project is just a way for you to gain the tools necessary for your next career step--an advanced degree, research techniques, soft skills, published papers, etc. My dissertation project studied whole organs, my friend’s dissertation project was on a single organelle in a cell--but we’re both now working at the same team at a very clinically-focused cancer research company that hired us not for dissertation projects but because of our PhDs in Cancer Biology.
And the fact that we both finished grad school with our mental health intact is a testament to choosing good PIs who fostered supportive lab environments. No matter how cool a project is, it’s not worth the sacrifice of your health. And grad school is a looooong time; some PhDs can take upwards of 6+ years. That’s a good chunk of time that can have lasting effects on the rest of your life.
In conclusion, to quickly summarize what I just said here, and in those links above:
Choose a field/program you’re passionate about (at least passionate enough to be willing to make it a career for the rest of your life)
Choose a grad school that makes you feel at home and has all the resources you need (eg. financial aid)
Choose a mentor and lab that supports you as a person
Choose a project that excites you and that you can grow from
And the rest will slowly fall into place :)
I hope that helps get you started. Let me know if you have any other questions! My private messaging is also open if you’d like a more 1-on-1 chat!
30 notes
·
View notes
Link
The U.S. War on Cancer, certainly a worthy cause, is a collection of programs stretching back more than 40 years and abiding under many banners. The latest is the Cancer Moonshot, launched in 2016 by then U.S. Vice President Joe Bidden, and passed as part of the 21st Century Cures Act. Over the years, computational technology has become an increasingly important component in all of these cancer-fighting programs, hand-in-glove with dramatic advances in understanding cancer biology and a profusion of new experimental technologies – DNA sequencing is just one.
Recently, deep learning has emerged as a powerful tool in the arsenal and is a driving force in the CANcer Distributed Learning Environment (CANDLE) project, a joint effort between the U.S. Department of Energy (DOE) and the National Cancer Institute (NCI), that is also part of the U.S. Exascale Computing Project (ECP). (For an overview, see HPCwire article, Enlisting Deep Learning in the War on Cancer).
Think of CANDLE as an effort to develop a computational framework replete with: a broad deep learning infrastructure able to run on leadership class computers and other substantial machines; a collection of very many deep learning tools including specific networks and training data sets for use in cancer research; and potentially leading to development of new therapeutics – drugs – for use against cancer. Currently, CANDLE helps tackle the specific cancer challenges of three cancer pilots, all using deep learning as the engine. Importantly, the three pilots have begun to yield results. The first software and data set releases took place in March, and just a few weeks ago, the first stable ‘CANDLE infrastructure’ was released.
Rick Stevens, ANL
In the thick of the CANDLE work is principal investigator Rick Stevens, also a co-PI of one of the pilots and associate laboratory director for the computing, environmental and life sciences directorate at Argonne National Laboratory (ANL). Stevens recently talked with HPCwire about CANDLE’s progress. The work encompasses all things deep learning – from novel network types, and algorithm development, to tuning leadership class hardware platforms (GPU and CPU) for deep learning use. Results are shared on GitHub in what is a roughly quarterly schedule.
Perhaps as important, “We are trying to plant inside the labs a critical mass of deep learning research people,” says Stevens. Just as CANDLE embodies broad hopes for applying deep learning to precision medicine, DOE has high hopes for developing deep learning tools able to run on leadership class machines and to be used in science research writ large.
“Everything is moving very fast right now,” says Stevens. “As the scientific community starts to get interesting results, two things will happen. One is we will be able to write papers and say ‘oh we took this network concept from Google and we applied it to material science and we changed it just this way and got this great result and now it is a standard way for designing materials.’ Those kinds of papers will happen.
“But then there is going to be another class of papers where I think in the labs, people that are interested in deep learning more generally and are interested in applying it to things other than driving a car or translating, but now are interested in applying it to particle physics or something – they are going to come up with some new deep learning algorithms, new network types, new layer types, new ways of doing inference for example, that are actually inspired by the problem in the scientific domain. That will take a bit longer.”
Clearly, there’s a lot going on here with CANDLE as the tip of the spear. Here’s a snapshot of the three pilots, which fall under Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) program underway:
RAS Molecular Project. This pilot (Molecular Level Pilot for RAS Structure and Dynamics in Cellular Membranes) is intended to develop new computational approaches supporting research already being done under the RAS Initiative. Ultimately the hope is to refine our understanding of the role of the RAS (gene family) and its associated signaling pathway in cancer and to identify new therapeutic targets uniquely present in RAS protein membrane signaling complexes.
Pre-Clinical Screening. This pilot (Cellular Level Pilot for Predictive Modeling for Pre-clinical Screening) will develop “machine learning, large-scale data and predictive models based on experimental biological data derived from patient-derived xenografts.” The idea is to create a feedback loop, where the experimental models inform the design of the computational models. These predictive models may point to new targets in cancer and help identify new treatments. Stevens is co-PI.
Population Models. This pilot (Population Level Pilot for Population Information Integration, Analysis and Modeling) seeks to develop a scalable framework for efficient abstraction, curation, integration and structuring of medical record information for cancer patients. Such an ‘engine’ would be enormously powerful in many aspects of healthcare (delivery, cost control, research, etc.).
The early work has been impressive. “In March, we put out a set of seven cancer deep learning benchmarks on GitHub that represent each of the three pilot areas. They represent five or six different deep learning network types and have self-contained data that you can download. That was a first major milestone. A couple of weeks ago we pushed out our first release of reliable CANDLE infrastructure. It contains what we call the supervisor which is infrastructure for running large scale hyperparameter searches (for network development) on the leadership computing platforms,” says Stevens.
Hyperparameters are things like the network structure (number and types of layers), learning parameters (activation functions, optimizers, learning rate) and loss functions. They are things that you set before training and are not learned during training. For any given problem there are many combinations of hyperparameters that are possible and often it is important to search for good combinations to get improved model performance.
“It runs here are Argonne, at NERSC and Oak Ridge National Laboratory, and [with some modest modification] runs pretty much on anybody’s big machines. What it does is manage hundreds or thousands of model runs that are trying to find goods choices of model hyperparameters.” The latter is indeed a major accomplishment. (Links to CANDLE releases so far: CANDLE Version 0.1 architecture: http://ift.tt/2vMgKTO; CANDLE benchmarks: http://ift.tt/2wEYFTR )
Already there have been surprises as illustrated here from Stevens’ pre-clinical screening project.
“We think we understand biology, and so we [were] thinking ‘we know relationships about the data and we can somehow structure the problem in a way that is going to make the biologically relevant information more accessible for the networks.’ The network surprised us by basically saying, ‘no that isn’t helping me, let me do it my own way’ in discovering the features that matter most on its own,” says Stevens.
“Of course I am anthropomorphizing but that was essentially the thought process that we went through. We were surprised by the fact that the machine could do better than we could. I think ultimately we’ll be right. It won the first round, but we’re going to get more clever in how we encode, and I think we’ll still be able to show that us working with the network will do a better job than either working alone.”
CANDLE INFRASTRUCTURE TAKES SHAPE As described by Stevens, CANDLE is a layered architecture. At the top is a workflow engine – a Swift/T-based framework called Extreme-scale Model Exploration with Swift (EMEWS) was selected – that manages large numbers of runs on the supercomputers and can be adapted for other more moderate size machines.
“We are using technology like NoSQL database technology for managing the high level database component, and these things are running on GPU-based systems and Intel KNL-based systems. We are trying out some of the new AMD GPUs (Radeon) and are experimenting with some more NDA technology,” Stevens says without discussing which systems have demonstrated the best performance so far.
“For the next level down in terms of scripting environment, we decided early on to build CANDLE on top of Keras, a high level scripting interface language for deep learning that comes out of Google. All of our models get initially implemented in Keras. All of our model management tools and libraries are also implemented in Keras. We’re doing some trial implementation directly in neon (Intel) and MXNet (Amazon),” says Stevens, but there are no plans to switch.
One of the things CANDLE leaders liked about Keras is it sits easily on top of all the major frameworks – Tensorflow, Theano, MXNet, and Microsoft’s Cognitive Toolkit (CNTK). Stevens says, “We don’t program typically directly in those frameworks unless we have to. We program in the abstraction layer on top (Keras). That gives us a lot of productivity. It also gives us a lot of independence from specific frameworks and that is really important because the frameworks are still moving quite quickly.”
Google’s TPU, an inference engine, has not been tested, but the “TPU2 infrastructure is on our list of things to evaluate.” Google, of course, has shown no inclination to sell TPU2 on the open market beyond providing access in the Google cloud (See HPCwire article, Google Debuts TPU v2 and will Add to Google Cloud).
“I think our CANDLE environment in principle could run in the Amazon cloud or Google cloud or Azure cloud, in which case if you were going to do that we would certainly target the TPU/GPU for acceleration. Right now our focus is in CANDLE because it is part of the ECP to target the DOE Exascale platforms as our first priority target. Our second priority target are other clusters that we have in the labs,” says Stevens. CANDLE also runs on NIH’s Biowulf cluster.
Stevens says the project has been talking to Intel about the Nervana platform and the Knights Mill platform and is, of course, already using Nvidia P100s and will work with the V100 (Volta).
OLCF’s recently deployed DGX-1 artificial intelligence supercomputer by NVIDIA, featuring eight NVIDIA Tesla GPUs and NVLink technology, will offer scientists and researchers new opportunities to delve into deep learning technologies.
A major governance review took place about a month ago and CANDLE received the go-ahead for second year. Stevens says there are now about 100 people working on CANDLE from four different labs and NCI. “I would say we have good collaborations going on with Nvidia, Intel, and AMD in terms of deep learning optimization interactions with architecture groups. We are also talking to groups at IBM and Cray, and we’ve had a big set of demonstration runs on the three big open science computers.”
PRE-CLINICAL DRUG SCREENING SURPRISE Of course the most immediate point of all this infrastructure work is to do worthwhile cancer research. Stevens is closest to the pre-clinical screening project whose premise is straight forward. Screen pairs of known drugs against cancerous cell lines and xenograft tumors, measure the effect – inhibition, growth, etc. – and build predictive models from the data. Most of the work so far has been done with cell lines grown in dishes but the effort to gather drug response data from human tumor grafts on rodents (human tumor xenograft model) is ramping up.
Says Stevens, “NCI took the 100 small molecule FDA cancer drugs and devised a high throughput screen that did all pairs of those drugs and concentration levels so we have something like 3.5 million experimental results there. We’ve been building a deep learning model that learns the relationship between the molecular properties of the tumor, in this case the cell line, and the structural properties of the pairs of drugs, and then predicts the percent growth inhibition.
“So we have millions of these pairs of drugs experiments and we have the score of how well the growth was inhibited. The machine learning model is trying to generalize from that training data, a representation that allows us to predict a result for a cell line and a drug combination that it hasn’t seen before. It’s a deep learning model. It’s a multi layered neural network and uses convolution.”
One surprise was how well a one-dimensional convolution layer neural network works on this problem.
“A priori we didn’t believe it would work, and it has better convergence than the fully connected network layers which was the way we thought we had to do it,” he says. “So that’s an interesting insight. The models are starting to become accurate enough and there is a lot of more work to do, but the initial results are encouraging. We’ve got models that are around 87 percent accurate in predicting whether the drug pair will outperform a single drug or not.
“The primary thing we are trying to do is get to a basic model architecture that has a high level of base line predictivity and then we can start optimizing it. Where we are now is encouraging. I think we can do better but we are showing that we have predictive power by building these models but we are also showing we need more data out of the tumors, not different types of data for the same tumor. We need more tumor samples from which we have highly curated molecular assay data and drug screening data,” he says.
There have already been some interesting results: “I have to be careful what I say because we are in the process of publishing those,” he says.
The RAS work is centered at Livermore. A well-known cancer promoter, the RAS family of proteins and pathways has been widely studied. It turns out ANL has the ability to synthesize “nanodiscs” with both wild type RAS and mutated RAS embedded in cell membrane and to obtain images and other biophysical measurements of the RAS proteins. That data is being used to train the simulations.
“The problem that team has been focusing on first is to reproduce computationally the phenomenon known as rafting – rafting is where more than one RAS protein aggregates [in the cell membrane] because that changes the dynamics of the signaling pathway. We have experimental data that shows there’s at least three different states of the membrane. What we are trying to do with the simulations is reproduce that basic biological behavior as a confidence builder exercise before we go to a more complex scenario,” says Stevens.
“One of the challenges is to extract the same kind of observational data from the simulation that you get from the experiment so one of the novel uses of machine learning has been to actually recognizing the different rafting states,” he says.
MAKING THE MODELS To a large extent the network training concepts are familiar. Train lower levels of the network to basic elements progressing to more specific assemblages of those elements at higher levels. Consider training a network to recognize cars, says Stevens. The lower network levels are trained on things like edges, shadows, colors, vertical and horizontal lines. The upper portion is trained to recognize things like windshields and hubcaps, etc. There’s lots of new and familiar tools to do this. Convoluted networks are hardly new. Generative networks are hot. He and his team are sorting through the toolkit and developing novel ones as well.
Returning to the auto recognizing analogy, he says “[If] now I want it to recognize a sailboat and different types of sailboats, the part of the network that recognized hubcaps and windshields isn’t going to be very useful but the part that learned how to recognize straight edges and colors and corners and shadows is essentially going to be the same. We think the same concept can apply to these drug models, or these drug response models. We are training the network to learn the difference between tumors and regular tissue, training it to recognize the difference between brain cancer and liver cancer, between colorectal cancer and lung cancer, for instance.
“They are recognizing what cancer is, the features that distinguish between normal cells and cancerous cells, and the features that relate to drug effectiveness. The models are trained on drugs, learning drug structures, learning the basic features of how molecules get put together – the kind of basic building blocks, side chains and rings and so on that are in molecules. Those don’t change. We are training on all the known drugs, not just cancer drugs, but all the known drugs, all the chemical compounds that are in the big libraries, so we learn basic chemistry at the bottom so we can combine the networks essentially to predict drug response. They don’t have to relearn most basic features. We just have to in some sense tune them up on the specific patient populations that you are looking for. I think these models could hop from preclinical screening application to actual use and clinical practice fairly quickly.”
FANNING THE DEEP LEARNING FLAME In some sense, deep learning, close kin to the more familiar data analytics, is a new frontier for much of science. Squeezing it under the HPC umbrella makes some people nervous. 64-bit double precision is nowhere in sight. Low precision doesn’t just work fine, it works better. How is this HPC, ask some observers in the HPC community?
Programing frameworks also present a challenge. Consider this snippet from a recent blog by Intel’s Pradeep Dubey, Intel Fellow and director of Intel’s Parallel Computing Lab, on the convergence of HPC and AI, “…unlike a traditional HPC programmer who is well-versed in low-level APIs for parallel and distributed programming, such as OpenMP or MPI, a typical data scientist who trains deep neural networks on a supercomputer is likely only familiar with high-level scripting-language based frameworks like Caffe or TensorFlow.”
Whatever your view of the computational technology being pressed into service, deep learning is spreading in the science community, and indeed, that is one of the CANDLE project’s goals. All of the major labs have deep learning programs and CANDLE has an aggressive ongoing outreach effort to share learnings.
“The CANDLE project is having a lot of impact [just] through its existence. We have kind of turned on a lot of people at the labs who say, ‘oh machine learning is real and deep learning is real and we are making sure that these future architectures we’re building now, the ones we are deploy in the 2021 timeframe are actually well-suited for running deep learning.’ That is causing the scientific community to say I should think about whether I could use deep learning in my problem,” says Stevens.
The post Deep Learning Thrives in Cancer Moonshot appeared first on HPCwire.
via Government – HPCwire
0 notes
Text
Running Hue on a Raspberry Pi Hadoop Cluster
Hadoopi - the Raspberry Pi Hadoop cluster
This project contains the configuration files and chef code to configure a cluster of five Raspberry Pi 3s as a working Hadoop running Hue.
This video shows how to set up and configure the cluster using this code.
youtube
The versions of installed Hadoop components are:
hadoop 2.6.4
hue 3.11.0
hbase 1.2.4
pig 0.12.1
hive 1.2.1
spark 1.6.2
livy 0.2.0
oozie 4.3.0
sqoop 1.99.4
solr 4.10.4
impala - not supported
Inspiration
Running Hadoop on Rasberry Pis is not a new idea, a lot of work has been done by individuals and I wanted to make sure their efforts were recognised. Their work and formed the basis of my attempts and inspired me to start the project.
Jamie Whitehorn - https://www.youtube.com/watch?v=n6ONKshWSMg
Jonas Widrikson - http://www.widriksson.com/raspberry-pi-2-hadoop-2-cluster/
Carsten Mönning - https://blogs.sap.com/2015/04/25/a-hadoop-data-lab-project-on-raspberry-pi-part-14/
Bram Vandewalle - http://www.becausewecangeek.com/building-a-raspberry-pi-hadoop-cluster-part-1/
DQYDJ - https://dqydj.com/raspberry-pi-hadoop-cluster-apache-spark-yarn/
Objectives
Learn
My day job uses the Cloudera distribution of Hadoop, which provides a great management interface, but that means I’m somewhat shielded from the inner workings of Hadoop configuration and tooling. I decided to put this distribution together to see if was feasible/practical to run Hadoop on a cluster of Raspberry Pis, but also to get more exposure to it’s tooling and configuration. At the same time I wanted a feature rich and easy to use “suite” of tools so I based the project around Hue. I also wanted it to be easily recreateable so used Chef.
Caveats
There are some caveates to bear in mind when using this tool.
No Impala - don’t think the Pi has enough power whilst running other Hadoop components - happy to receive pull requests :-)
No Sentry - access to the data on the cluster will be based around HDFS security
Not a production reference - it’s a learning exercise, given the performance of the cluster you really wouldn’t want to run anything “real” on there.
Teeny amount of memory so only basic stuff - The 1GB of ram on the Pi is a real limitation, you’re really only going to be able to one task at a time, so go easy on it
Its slowwwwwwwww - the combination of teeny amount of RAM, 4 cores and wifi networking means this is built for speed, be realistic with your expectations!
Setup requires basic linux command line fu and understanding of network configuration - you are going to be compiling applications, running chef code and possibly doing a little fault finding, you’ll need some basic linux sysadmin skills for this.
It’s compiles and configures correctly as of NOW - there are loads of dependencies on 3rd party libraries and packages, both when compiling binaries or configuring the cluster, but things get move, deleted or just change, you’ll need be able to diagnose problems and change settings.
Ask if you want help - github issues are ideal but please be clear in what attempts you have made to diagnose and fix.
The hardware
To build the cluster you are going to need:
5 x Raspberry Pi 3s
Nylon Spacers to stack the Pis
Acrylic Raspberry Pi case for base and lid to keep the dust off
Wireless Router - I’m using a TP-Link tl-wr802n travel router
Anker 60w 6 usb port hub
6 usb cables
5 x Samsung Evo+64GB micro sd cards
Computer for administering via ssh, running a webserver and a web browser to access Hue
Just a quick note on the micro sd cards, perfromance of cards can vary wildly, I was recommended the Evo+ cards as their performance at reading and writing small files was very good, you can check the performance of yours using https://github.com/geerlingguy/raspberry-pi-dramble/blob/master/setup/benchmarks/microsd-benchmarks.sh
Making Binaries
Although most of hadoop eco system is written in java and available in binary format, there are some components that need to be built for the target architecture for performance. As the Raspberry Pi is ARM based we simply can’t take the precompiled binaries and run them, we’re going to have to compile those. These are Hadoop (with the correct version of protobuf libraries that we will also need to build), Oozie and Hue.
Install the Rasbian Jessie Lite version dated 23/09/16 onto you sd card, the development of the project was based around this version.
http://downloads.raspberrypi.org/raspbian/images/raspbian-2016-09-28/
This version has a few “features” that are really useful, firstly it will auto expand the file system on first boot, secondly the SSH server is enabled by default (which is probably an error) but it means we can configure the Pi headless without the need of doing basic service config without the fuss of connecting a keyboard and monitor.
Compile protobuf
Locate you Pi’s ip address (via some form of network scan, or go through the fuss of connect a monitor and keyboard) then ssh to it as the pi user. then download and unpack the protobuf v2.5 source, tweak the build script so that it refers to the new location of the google test suite and build and install the libraries:
sudo -i apt-get update apt-get install dh-autoreconf wget https://github.com/google/protobuf/archive/v2.5.0.zip unzip v2.5.0.zip cd protobuf-2.5.0 vi autogen.sh
Change the references to the google test suite:
# Check that gtest is present. Usually it is already there since the # directory is set up as an SVN external. if test ! -e gtest; then echo "Google Test not present. Fetching gtest-1.5.0 from the web..." wget https://github.com/google/googletest/archive/release-1.5.0.zip unzip release-1.5.0.zip mv googletest-release-1.5.0 gtest fi
Then generate the build configuration files
./autogen.sh ./configure --prefix=/usr
Build and install protobuf
make make check make install
Compile Hadoop
We now need to compile the Hadoop binaries, download and unpack the Hadoop 2.6.4 source, tweak pom.xml so it bypasses the documentation generation as this fails on the Pi, apply the HADOOP-9320 patch and build the binary.
cd apt-get install oracle-java8-jdk wget http://apache.mirror.anlx.net/hadoop/common/hadoop-2.6.4/hadoop-2.6.4-src.tar.gz tar -zxvf hadoop-2.6.4-src.tar.gz vi hadoop-2.6.4-src/pom.xml
Disable the problem step in by adding the following to <properties>…</properties>
<additionalparam>-Xdoclint:none</additionalparam>
Next apply the HADOOP-9320 patch
cd hadoop-2.6.4-src/hadoop-common-project/hadoop-common/src wget https://issues.apache.org/jira/secure/attachment/12570212/HADOOP-9320.patch patch < HADOOP-9320.patch cd ~/hadoop-2.6.4-src/
Next install a whole bunch of build tools and libraries:
apt-get install maven build-essential autoconf automake libtool cmake zlib1g-dev pkg-config libssl-dev libfuse-dev libsnappy-dev libsnappy-java libbz2-dev
Then run the build using maven
sudo mvn package -Pdist,native -DskipTests -Dtar
Once build package the archive ready for deployment
cd hadoop-dist/target/ cp -R hadoop-2.6.4 /opt/hadoop-2.6.4 cd /opt tar -zcvf /root/hadoop-2.6.4.armf.tar.gz hadoop-2.6.4
Compile Hue
Download the Hue 3.11.0 source, unpack it, apply the necessary patches and tweak the spark_shell.py so it defaults to 256MB for spark driver and executor memory rather than 1GB.
Install the build dependencies:
apt-get install python-dev libsasl2-dev libxml2-dev libxslt-dev libkrb5-dev libffi-dev libldap2-dev libmysqlclient-dev libsqlite3-dev libgmp3-dev libssl-dev
Download and unpack the Hue source
cd /opt wget https://dl.dropboxusercontent.com/u/730827/hue/releases/3.11.0/hue-3.11.0.tgz tar -zxvf hue-3.11.0.tgz cd hue-3.11.0
Download the patch to fix the example loading issues with 3.11.0, patch the desktop/core/src/desktop/api2.py and desktop/core/src/desktop/tests_doc2.py files:
wget https://github.com/cloudera/hue/commit/b059cec5c55737af3ceeb3a8cb2c0ce4e4d94b4d.patch patch < b059cec5c55737af3ceeb3a8cb2c0ce4e4d94b4d.patch
Change the driverMemory and executorMemory defaults from 1GB to 256MB in desktop/libs/notebook/src/notebook/connectors/spark_shell.py
Then build the apps:
make apps
Then package them up:
cd /opt tar -zcvf /root/hue-3.11.0.armf.tar.gz hue-3.11.0
Compiling Oozie
Finally we need to compile oozie. Download the oozie 4.3.10 source and unpack it, we need to change the target java version to 1.8 in the pom.xml file, then build the binaries, download the ext-2.2. library (to enable the oozie web interface) and copy to the applicable folder and package the files.
Download the files and unpack:
cd apt-get install oracle-java8-jdk maven wget http://archive.apache.org/dist/oozie/4.3.0/oozie-4.3.0.tar.gz tar -zxvf oozie-4.3.0.tar.gz cd oozie-4.3.0/
Change the target java version to 1.8 in pom.xml, then set a few environment variables to stop the build process running out of memory, and build the binary:
export MAVEN_OPTS='-Xmx512m -XX:MaxPermSize=128m' bin/mkdistro.sh -DskipTests -Puber
Now download the ext-2.2 library and package the files:
cd /root/oozie-4.3.0/distro/target/oozie-4.3.0-distro/oozie-4.3.0 mkdir libext cd libext wget http://archive.cloudera.com/gplextras/misc/ext-2.2.zip cd /root/oozie-4.3.0/distro/target/oozie-4.3.0-distro/ tar -zcvf /root/oozie-4.3.0.armf.tar.gz oozie-4.3.0
Making the Compiled Files Available
Transfer the compiled binary files to you computer and start the python webserver to make them available to the Pis as you configure the cluster:
python -m SimpleHTTPServer
Installing and Configuring the Cluster
The cluster will comprise of 5 Raspberry Pis, 3 of them will be configured as worker nodes (worker01, worker02 and worker03) and have the HDFS Datnode and Yarn Nodemanager Hadoop components installed. The remaining two Pis will be setup as master01 and master02.
The master nodes will have the the Hadoop components:
master01
HDFS Namenode
Yarn Resource Manager
Hue
Hive
Pig
Oozie
Sqoop
master02
Hbase
Zookeeper
Spark & Livy
Solr
Networking
The cluster is setup to run on the 10.0.0.x network range:
10.0.0.11 - master01
10.0.0.12 - master02
10.0.0.21 - worker01
10.0.0.22 - worker02
10.0.0.23 - worker03
10.0.0.9 - administering computer running python webserver
The TP-Link router is setup in “WISP Client” mode so it bridges the two wifi networks from 10.0.0.x to 192.168.2.x that way I can set the cluster up so it has outbound internet connectivity from the 10.0.0.x network on via my 192.168.2.x network. The great advantage with this setup is the cluster and router can taken away from my 192.168.2.x network and use the cluster without having to reconfigure the network.
You can modify the network configuration by modifying the chef attributes or refactoring the network recipe. You may also want to add the nodes to your administering computer /etc/hosts file.
Code
The code for the projects is available over at https://github.com/andyburgin/hadoopi
Installing
Firstly write the Jessie Lite version dated 23/09/16 onto your sd cards. We will setup the Pis in turn, worker01-03 and then master02, for the final step when we setup master01 which will need all the Pi’s powered on as it modifies files on the HDFS file system. This means you’ll only need on pi connected to ethernet.
So insert the first sd card into one of your Pis, connect the ethernet cable and power it up, wait a minute whilst the Pi boots and SSH into the pi, you can determine the ip address of your pi by either using a network scanner or by connecting a monitor to the hdmi port.
Once we have ssh'ed into the Pi we need to update the system:
sudo -i apt-get update
Installed git and chef:
DEBIAN_FRONTEND=noninteractive apt-get -y install chef git
Clone the code from github to the Pi:
git clone https://github.com/andyburgin/hadoopi.git cd hadoopi
Set your wifi SSID and password as environment variables:
export WIRESSID=myssid export WIREPASS=mypassword
Then run chef against the worker01.json file
chef-solo -c solo.rb -j worker01.json
Wait for it to finish and:
poweroff
You now need to repeat these steps using a fresh sd card for worker02, worker03 and master02.
For the setup of master01 place the final sd card in the pi with the ethernet connection, setup the remaining 4 pis with the previously configured sd cards and power up all of the pis. As before ssh into the Pi you are configuring, update the system, install git & chef, clone the code, set you network SSID & password, then run:
chef-solo -c solo.rb -j master01.json
Then we need an extra chef run to install additional Hadoop components and configure files on the HDFS filesystem that runs across the cluster:
chef-solo -c solo.rb -j master01-services.json
Then ssh into all of the nodes (via their 10.0.0.x ip addresses):
poweroff
Starting and Stopping the Cluster and Installing Examples
As a one off exercise before we start the cluster lets install some test data into mysql for later testing with sqoop, we do this before we start the Hadoop services as this will require too many system resources to tun both. SSH to master01 where our mysql instance runs (this is required to hold Hue configuration data, but we can also use it to hold test data too) and run:
sudo -i git clone https://github.com/datacharmer/test_db.git cd test_db mysql < employees.sql mysql -u root -e "CREATE USER 'hduser'@'%' IDENTIFIED BY 'hduser';" mysql -u root -e "GRANT ALL PRIVILEGES on employees.* to 'hduser'@'%' WITH GRANT OPTION;" mysql -u root -e "FLUSH PRIVILEGES;"
Starting the Cluster
On master01 and master02 you will find some scripts in the /root/hadoopi/scripts folder. After powering on the cluster (you no longer need the ethernet connection as all nodes will connect to the wifi connection) start the cluster on master01 by issuing:
sudo -i cd ~/hadoopi/scripts ./master01-startup.sh
Then on master02 run:
sudo -i cd ~/hadoopi/scripts ./master02-startup.sh
Once those scripts have sucessfully run you are now ready to configure Hue.
Configuring Hue
We access hue via a web browser, point you we bbrowser your web browser at:
http://master01:8888/
The first time you hit the url you’ll be asked to create a Hue administrator user, the cluster is configured to work withe the “hduser” user, so make sure you use that with a password of your choice.
From the “Quick Start Wizard” select the “examples” tab and one by one install each of the examples. Frome the “Data Browser” menu take a look at the “Metastore Tables” to check Hive data is available and the “HBase” option to view the data there. Finally select one of the Dashboards under the “Search” menu to heck the Solr indexes are available.
Stopping the Cluster
Powering down the cluster is as easy as starting it, on master01 run:
sudo -i cd ~/hadoopi/scripts ./master01-stop.sh
Then on master02 run:
sudo -i cd ~/hadoopi/scripts ./master02-stop.sh poweroff
Finally on each of the worker nodes run:
sudo poweroff
From now on you can simply power on the cluster and run the startup scripts on master01 and master02 as root. Once you have finished using the cluster shut it down by running the stop script on master01 and master02, then running the “poweroff” command on each of the 5 nodes as root.
Hue Examples
I won’t go through every example using installed in hue, watching the vide will convey far more, but I’ll cover some of the key ones and allow you to explore. Please remember the Raspberry Pi has a teeny amount of memory and runs things very slowly, be patient and don’t submit more than one job at once, doing so will more than likely cause your jobs to fail.
Hive Examples
From the “Query Editor” menu select “Hive”, in the resulting query window select one of the example scripts e.g. “Sample Salary Growth” and hit the “play” button to submit the query.
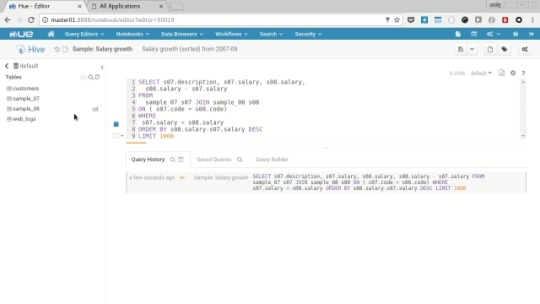
Choose the “Job Browser” icon and open in a new tab, watch the job transition from Accepted->Running->Succeeded.
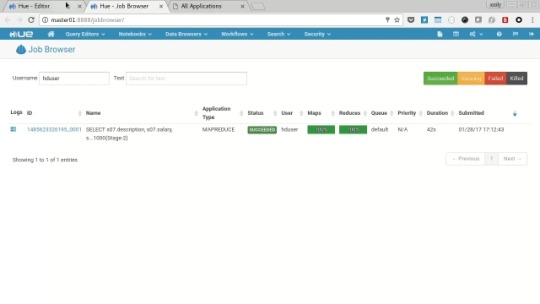
Return to the original query tab and you’ll see the results displayed in the browser.
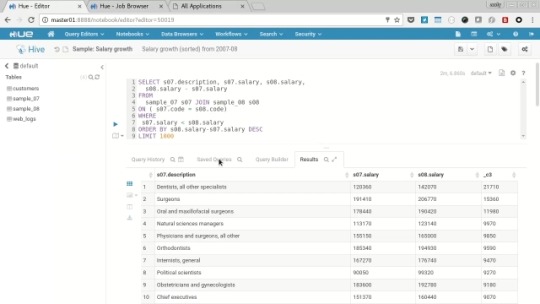
Pig Example
From the “Query Editor” menu select “Pig”, then select the “samples” tab and then the “Upper Text (example)”, in the resulting query editor amend the 2nd line to:
upper_case = FOREACH data GENERATE UPPER(text);
Click the play button to submit the query, you’ll be prompted to specify an output path, so choose “/tmp/pigout” and click “Yes”.
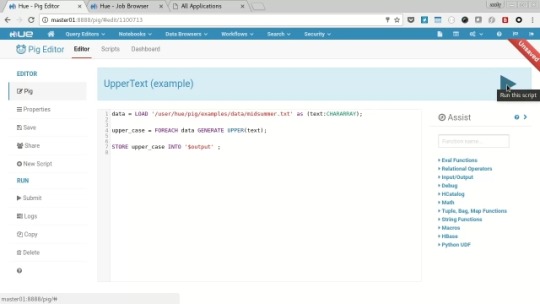
Go to the job browser and wait for the two generated jobs to finish, then click the “HDFS Browser” icon and navigate to /tmp/pigout and view on e of the “part-m-????” file and see the generated text.
Spark Notebooks
Spark notebooks are one of the best features of Hue, allowing you to edit code directly in the browser and run it via spark on the cluster. We’ll try out the three supported languages Python, Scala and R (we don’t have Impala support).
Select “Notebooks” menu and open the “Sample Notebook”, we’ll need to start a session for each language, we’ll try Python first so click the “context” icon to open the context menu and then choose “Recreate” next to the PySpark option, this creates a Spark job viewable via the job browser.
When the session is ready and the job is running, click the “play” icon next to each of the 4 examples and the cluster will calculate the result and display it in the browser, feel free to experiment with the results and edit the code.
When you have finished with the PySpark examples you need to kill the session/job, so on the context menu click “close” next to the PySpark menu.
To try the Scala examples click “Recreate” next to the Scala option on the context menu to create the session and associated job. When running you’ll be able to edit Scala code in the browser and interact with the results.
After trying both examples kill the session/job by closing the session on the context menu.
Skip over the Impala examples as they aren’t supported.
Start the R session by clicking “create” next to the option and when the job/session have started edit the path in the R sample to /tmp/web_logs_1.csv The R example doesn’t used HDFS so you’ll need to SSH onto each worker and run:
curl https://raw.githubusercontent.com/cloudera/hue/master/apps/beeswax/data/web_logs_1.csv -o /tmp/web_logs_1.csv
Back on the Notebook click the play icon next to the code and examine the output including plot.
Finally close the session on the context menu so the spark job ends.
Sqoop example
We’re going to use Sqoop to transfer the employee sample data we installed earlier into HDFS. To do this we’ll need to configure two Sqoop links and a Sqoop Job.
Create the first link by selecting the “Data Browser” and then “Sqoop”. Choose “Manage Links” then “Click here to add one”
Name: mysql employees in
Connector: generic-jdbc-connector
JDBC Driver Class: com.mysql.jdbc.Driver
JDBC Connection String: mysql://master01/employees
Username: hduser
Password: ******
Then click “Save”, choose “Manage Links” again to add the second link. Click “New Link” and enter the follwoing values:
Name: hdfs out
Connector: hdfs-connector
JDBC Driver Class:
JDBC Connection String:
Username: hduser
Password: ******
We need to then edit that link to complete setting it up, click “Manage Links” the the “hdfs out” link to bring up the edit page. Make the follwoing settings:
Name: hdfs out
HDFS URI: hdfs://master01:54310/
Finally click Save.
Next we need to configure the job, select “Click here to add one” and fill out the following:
Step 1: Information
Name: import employees from mysql to hdfs
From Link: mysql employees in
To Link: hdfs out
Step 2: From
Schema name: employees
Table name: employees
Table SQL statement:
Table column names:
Partition column name:
Null value allowed for the partition column:
Boundary query:
Step 3: To
Output format: TEXT_FILE
Compression format: NONE
Custom compression format:
Output directory: /tmp/sqoopout
Start the job by clicking “Save and run”, navigate to the job browser and wait for the job to be submitted and run sucessfully. Then Navigate to hdfs browser and check the data in “/tmp/sqoopout”
Ozzie Examples
There are many Ozzie examples installed, so I’ll only talk about a few of them here. Lets firstly run the Shell example. Select “Query Editors” from the menu and the “Job Designer”, next click the “Shell” example, you’ll be presented with the job editor. I for scroll down you’ll see a parameter to the “hello.py” command is “World!”.
Click the “Submit” button (then confirm) and you’ll be presented with the workflow view of the job, wait for the job to run and and select the log icon next “hello.py”. In the resulting log you’ll see the phrase “Hello World!”
Next let’s look at amore complicated workflow, from the menu select “Workflows” -> “Editors” -> “Workflows”, on the resulting page select “Spark” and you’ll be presented with the job configuration page, for the spark workflow to run we will need to configure the settings for spark, click the “pen” icon to edit the job, next click the “cogs” icon on the spark step to access the steps properties. In the “Options List” filed enter:
--conf spark.testing.reservedMemory="64000000" --conf spark.testing.memory="128000000" --executor-memory 256m --driver-memory 256m --num-executors 1
Click the “disk” icon to save the changs to the workflow and click the “play” icon to submit the job, enter “/tmp/sparkout” into the output field and hit “Submit”. Again you’ll be presented with the workflow view, wait for the job to finish and then use the hdfs browser to navigate to the “/tmp/sparkout” folder and view one of the data files to check the spark job copied the intended files.
Solr Dashboards
Select “Search” from the menu and then each of the dashboards in turn (Twitter, Yelp Reviews and Web Logs).
You’ll see each present a variety of charts, graphs, maps and text along with interactive filtering features to explore the data held in the solr search indexes.
Reminder, Power Off the Cluster
Once you have finished experimenting with the cluster please don’t just turn off the power, you rick corrupting your data and possibly the sd card. So follow the procedure of running the shutdown scripts on master01 and master02, then running the poweroff command on each of the pis before turning the power off.
Wrapup
I hope you have fun setting up and playing with the code, I learned a tonne of stuff setting it up and I hope you do too. If you have any improvements then please send pull requests to github, any problems and I'll respond to issues there too.
0 notes
Note
I remember a while back you had a few posts about applying for the f31. Do you have any tips or resources that helped you out? I couldn’t find any tags on your blog about grants. Thanks so much!!
hi!
so after perusing through my swamp-of-a-blog i couldn’t find any posts i had made that specifically talked about the F31 (other than me complaining about it). Or maybe I did on certain aspects but not the whole thing… anyway! so i’m going to make one! just for you anon <3
Here’s the website for NIH’s Application Guide, which includes rules on formatting, how to submit, etc.
If you scroll down to the orange “F” for “Fellowship Instructions”, there are two PDF instruction manuals based on your planned date of submission (either before Jan 24, 2018 or after Jan 25, 2018, bc they updated a few things).
Here’s the PDF instruction manual for after Jan 25. Read through everything. I highly recommend making a summary table that lists all the documents you’ll need to write, a short description of what it is, the page limit, who writes it (you, you & PI, PI, co-sponsor, etc), and what page in the instruction manual you can go to for more info.
Make sure to work closely with your PI, your co-sponsor, and your university grants office. Please note that you do not submit the grant; your university grants office submits it on your behalf. Also in my case, they created the budget sheet. I recommend getting in touch with a rep from the grants office asap, because the internal due date is earlier than the NIH due date (bc the office needs to put everything together, etc). For example, my F31 submission date to my grants office was Dec 4, for the NIH due date of Dec 8.
Your PI will also need to work on some aspects of the grant with you (such as the Training Plan). Be sure to figure out a battle plan with draft deadlines with them. Same thing with your co-sponsor (if you have one, and it is highly recommended).
Letters of rec are also required, so be sure to give your writers at least a month to work on these. You may need to provide an outline of what you’d like them to focus on.
Many aspects of the F31 can come from “boilerplates” forms. These are generic things from your lab that can just be copy&pasted from previous applications, such as descriptions of Equipment, Institutional Environment & Commitment to Training, etc, though I recommend updating and customizing them as you see fit. Check with your PI or program coordinator to see if these are available.
I recommend finding an example of a funded F31 from a previous lab or program member, and noting the strengths in their application. Keep in mind format and content may have been updated for the current instructions, but some things are still the same. Also it always helps to see what other people have done when tackling something new.
The entirety of a good F31 will take a few months to write, edit, and finalize (assuming you’ve already done all your culling of scientific info, either experimentally or through literature searches). A semester to one year is what I’ve seen people spend on them. So start early! I recommend setting mini deadlines for yourself, and also scheduling at least an hour or two one day a week just to write/edit, and treat it like a class. Don’t trust yourself to “find the motivation”; discipline is what counts here.
What makes a good F31:
Scientifically sound and doable research project. Scientifically sound is self-explanatory (like having all the appropriate controls, etc), but also make sure your project is doable for a PhD student. “Boil the ocean” projects are never funded.
A well-rounded and productive support system. For example, your training goals and your PI’s training plan match, your PI and co-sponsor(s) have productive publication and grant records, your institutions has the necessary facilities and training programs for you to carry out your research and training plan, etc. Be sure to also think about things like career development, grant and manuscript writing workshops, etc.
No contradictions. Meaning you actually worked with your PI and co-sponsor on the grant, because if your PI’s training plan says you’ll be done in 4 years, but your training goal says 5, then that’ll raise eyebrows and indicate a lack of involvement or communication between you and your support system.
Clear, easy to understand writing. Edit edit edit edit edit! Good writing goes a long way. The easier you make the lives of your reviewers, the happier they’ll be, and the happier they are, the more generous their scores. Also don’t make your figure legends/text too small. Keep in mind that a lot of these grant reviewers are older, so anything smaller than 8-pt font may be too hard to read.
A little something extra. F31 reviewers want to see that should you be funded, you’ll get to do something above and beyond what your current stipend allows you to do. For example, visit and train with a collaborator in another part of the country.
Keep in mind that your F31 proposed project does not necessary have to be your dissertation project to the dot. The scientific community understands that research trajectories change, and no one’s going to check when you graduate if your specific aims are still the same. A previous PhD student in my lab proposed to study ginger in her F31 (which got funded), but her dissertation project had nothing to do with ginger (she switched focus to turmeric).
Also, don’t do this alone. Your PI and co-sponsor(s) should help you, and so should your program. See what resources they may have first before tackling this project. I also recommend taking a grant writing course (if possible) before doing this, as there are a lot of nitty gritty details that go specifically into grant writing.
And lastly, a word from someone who’s gone through the process: It’s gonna suck. There will be a point where you’ll get so sick of editing the same paragraph over and over, and every fiber of your being is going to scream for this to be over. But keep at it. Just do one word at a time, one sentence at a time. And sooner or later you will finish, and gain these lovely experience points and feel good about accomplishing such a huge challenge!
Best of luck! If you have any questions, please don’t hesitate to come talk to me. Even if it’s to vent (which I’ve done many-a times during this hellish process haha)!
14 notes
·
View notes