#mdn1!!!!!!
Explore tagged Tumblr posts
Note
may i request some more nsfw love n deepspace <3 i love the headcanons u posted aaa
one more—
tags: 18(+), mdni, very nsfw, suggestive content, fem!reader, short & sweet, men of LaDS!!
creator notes: omg i’m glad you liked them!! i haven’t played the game (i have but like when it first came out!) so i know none of the lore and hope i wasn’t writing the men ooc or anything aaaaa!!!! but i’m glad you enjoyed them enough to request for more!! thanks for requesting as well!

SYLUS—
actually makes you wear a collar and a leash
your first time together was rough, both of you fighting for dominance over each other but ultimately he won
degrades you!!!
calls you “kitten” when you please him but any other time he’s telling you how pathetic you are for wanting him as badly as you do
loves it when you’re a brat solely so he can break you later
is so into tying you up, forcing you to take whatever he gives you
especially likes it when he ties your hands behind your back and makes you ride him
loves to watch you struggle
you have a safe word for a reason!!!!!!
is 8 inches and thick as all hell, deep red tip, veiny
RAFAYEL—
the first time you both wanted to be intimate he was a nervous wreck
took you a few times to steady his hands and convince him that you wanted it too
worships you like a goddess when you fuck
i secretly think he has amazing stamina but hides it from you
that way you’re on round 3, overstimulated and he’s asking “once more please one more time my love.”
begs you for it!!!
i’d like to think he’s a switch! let’s you start things, get him all heated and needy, then he takes total control
also think he’s always in the mood
24/7 he wants to touch you, taste you but holds himself back and instead makes jokes
is probably 6.5 inches, curved, and flushed pink
XAVIER—
you two struggle to go on missions together anymore solely because you always want to touch each other
always requests for you to be his partner too
loves having sex under the stars
especially when you ride him and he gets to see the stars halo your head
his max number of rounds he’s gone without passing out is 3
once he wakes up though he’s ready to continue
knows every single one of your weaknesses
he rarely makes noise when you two fuck but constantly reminds you how good you look
“baby you look so beautiful.” he says as he has your legs on his shoulder, pounding into you
is roughly 7 inches, thicker at his base, a nice red tip
ZAYNE—
your first time with him was definitely either in his car, office, or his apartment
uses his tie to keep you quiet
he likes to let you think you’re in control but really you both know he is
uses the excuse that he’s “always cold.” and needs you to warm him up
forces you to look at him when he fucks you
especially when you want something, makes you look him in the eye as you beg him for it
praises praises praises!!!!!
“good girl.”
i also think he’s horny ALL THE TIME but only when you two are together!! then he stares at every move you make and thinks to himself how he’d take you right then and there
8.2 inches, veiny, thicker head that’s a pale pink
#zevrra zevrra!#zevrra’s hc’s#zevrra replies#add a lil spice 🌶️#mdn1!!!!!!#anon request#love and deepspace#love & deepspace#love and deepspace sylus#zayne love and deepspace#rafayel love and deepspace#xavier love and deepspace#sylus x reader#xavier x reader#zayne x reader#rafayel x reader#fem!reader#f!reader#lads#lads x reader#also so so so sorry but Sylus doesn’t give aftercare istg LSHSKSHLSH#anyway—
415 notes
·
View notes
Text
21+ please, guys.
Not all of the content is Safuhwerk
0 notes
Text
"And they were roommates" (teaser)
Eddie Munson x Onlyfans Reader
MDN1 18+
WC: 700
Summary: Eddie's crush on his roommate is constantly weighing him down to the point that he's desperate to find any content that reminds him of her so he can jerk off and go to sleep. Imagine his surprise when he finds a video of you, legs spread as you touch yourself proudly on camera
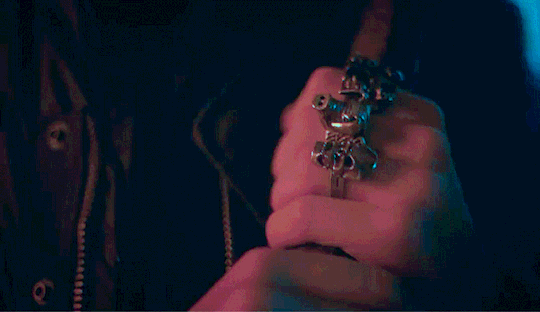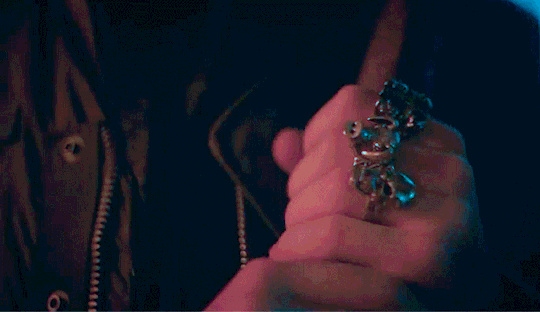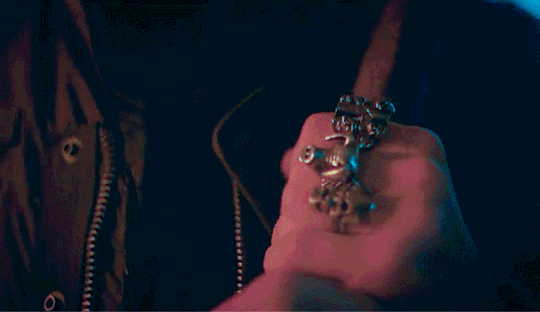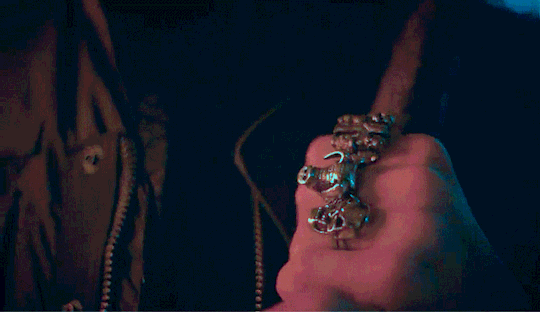
Eddie shouldn't be doing this.
Eddie knows he shouldn't be doing this.
It was an accident, a complete accident. Eddie has been secretly crushing on his roommate for a few months now but hasn't done or said anything about it yet. You weren't trying to kill him, you'd just come into the kitchen to grab a snack wearing a baggy T-shirt and some torturously small sleep shorts. You weren't even doing anything intentionally sexual to set him off, but it was enough to make Eddie excuse himself to bed early to get rid of his growing hard on.
He had touched himself to the thought of you, multiple times actually. But this was the first time he was looking up someone like you to help fuel his imagination. He was looking up your hair color, your body type stuff like that into his porn searches but wasn't actually expecting to find you. He must've been seeing things there was no way that it was actually you. The thumbnail had you in nothing but your bra, legs spread, hand in between your thighs as you touched yourself, proudly smiling into the camera.
Holy shit, He tapped on the video to make sure that he wasn't hallucinating. He couldn't help it. He didn't even bother loosening his jeans before shoving his hand into his underwear. The woman that he's been pining after for months is right there, on his phone screen, getting herself off in her room. Her room. Which shared a wall to his room in their cramped apartment. Eddie has had heart eyes for you the second you moved in. His friends knew about this, saying that his crush on you was painfully obvious. He just hoped that it wasn't obvious to you.
He actually struggled to speak to you for the first few days, until you and some mutual friends all went out for dinner. Steve eventually pulled him aside and threatened to embarrass him in front of you, as a way of forcing his confidence. It didn't take long to break the ice, discovering that you both had a lot of similar interests. Now, both you and Eddie feel safe to call each other pretty close friends. Watching horror movies together on the couch, smoking weed together while blasting music. You had even gone to see a few of his shows at the hideout when you weren't busy.
For now, Eddie continued pumping his leaking cock, trying to match your pacing to you through the screen of his phone. He's almost hypnotized watching your fingers disappear inside yourself wishing that it was his instead making you feel so good. He can see how wet you are from the glistening on your fingers when you pull them out, And the wet sounds it makes when you put your fingers back in. Fucking hell. The regret will sit heavy on Eddie's chest tonight, but all he can think about is how sweet you look whilst you continue sliding your fingers through your folds, whimpering softly against the pillow, trying to stay quiet. If only Eddie could be there, on his knees with his tongue between your legs whilst you slide your fingers into his curls. You probably tasted so sweet.
Eddie cums in his pants with a soft whimper. It was uncomfortable and desperate, the worst kind of dampness. He instantly cursed himself for not removing any of his clothing before wrapping his fist around his cock. In his defense, his discovery was sudden and exciting, and Eddie didn't even think about locking his bedroom door, let alone preparing himself properly. As the video continues playing he starts scrolling through your channel and is surprised not just by the amount of videos you've posted. But the views, the likes, the comments, there were just so many. Not just on this one but all of them.
You'd never really told Eddie what you do for a living, it never really came up in conversation. He only knew that you work from home, which technically isn't a lie. But this is never what he would've guessed what you meant. The video eventually ended, fading to black with some white text appearing. Eddie enlarged the video again to read it. ‘Hey Guys!!! Thank You So Much For Watching! (˶ᵔ ᵕ ᵔ˶) To see more of me Check Out My OnlyFans!!! Link Here!’
He was fucked
A/N: this is just a little taste of the first chapter of this fic ;) rn the word count just hit 7k but didnt want to post something unfinished. I'm touching up the ending and don't know how long it will take me to complete it. Hope you enjoyed this little teaser 😋
#my fic#and they were roommates#eddie x reader#eddie smut#eddie munson#eddie#eddie stranger things#eddie munson smut#eddie munson imagine#eddie munson x reader#eddie munson headcanon#stranger things s4#stranger things smut#stranger things fanfiction#stranger things#eddie munson x fem!reader#eddie x fem!reader#eddie munson x fem!reader fluff#teaser#modern eddie munson#modern au#eddie munson fan fiction#eddie munson x reader smut#eddie munson x y/n#eddie munson x you#eddie munson x female reader#eddie munson x fem!reader smut
1K notes
·
View notes
Text
Data Management and Visualization: Week 4 Assignment
Creating your own graphs :SAS and Python
SAS: Code




Output:



















Python: Code
#importing Pandas and NumPy libraries import pandas import numpy import seaborn import matplotlib.pyplot as plt #Loading the Mars Crater dataset DATA = pandas.read_csv('marscrater_pds.csv',low_memory=False) pandas.set_option('display.max_columns',None) pandas.set_option('display.max_rows',None) #Printing the number of rows and coloums in the dataset print(len(DATA)) print(len(DATA.columns)) #Converting the value of the object into numeric DATA['LATITUDE_CIRCLE_IMAGE']= pandas.to_numeric(DATA['LATITUDE_CIRCLE_IMAGE']) #defining a new variable in order to divide the latitude into three groups, viz >25, <-25, and -25<0<25 def LAT (row): if row['LATITUDE_CIRCLE_IMAGE'] >25: return 1 if row['LATITUDE_CIRCLE_IMAGE'] < -25: return 2 else: return 0 DATA['LAT']= DATA.apply(lambda row:LAT(row),axis=1) #Printing the number of rows and coloums in the dataset print(len(DATA)) print(len(DATA.columns)) #subsetting to obtain the specific data required ie the data coming in the range of latitude -25 to +25 EQTR=DATA[(DATA['LAT']==0)] EQTR2=EQTR.copy() #Printing the number of rows and coloums in the dataset print(len(EQTR2)) print(len(EQTR2.columns)) #Converting the value of the object into numeric EQTR2['DIAM_CIRCLE_IMAGE']= pandas.to_numeric(EQTR2['DIAM_CIRCLE_IMAGE']) EQTR2['DEPTH_RIMFLOOR_TOPOG']= pandas.to_numeric(EQTR2['DEPTH_RIMFLOOR_TOPOG']) EQTR2['NUMBER_LAYERS']= pandas.to_numeric(EQTR2['NUMBER_LAYERS']) #Omitting missing data EQTR2['DEPTH_RIMFLOOR_TOPOG']=EQTR['DEPTH_RIMFLOOR_TOPOG'].replace(0, numpy.nan) EQTR2['NUMBER_LAYERS']=EQTR['NUMBER_LAYERS'].replace(0, numpy.nan) #Data management of latitude variable along with frequency and precentages EQTR2['EQR']= pandas.cut(EQTR2.LATITUDE_CIRCLE_IMAGE,[-25,-15,-5,5,15,25]) EQTR2['EQR']=EQTR2['EQR'].astype('category') print("Latitude grouped") FREQ1=EQTR2['EQR'].value_counts(sort=True) print(FREQ1) PER1=EQTR2['EQR'].value_counts(sort=True, normalize=True)*100 print(PER1) #Data management of diameter variable along with frequency and precentages EQTR2['DIA']= pandas.cut(EQTR2.DIAM_CIRCLE_IMAGE,[1,1.5,2.5,3.5,4.5,1096.65]) EQTR2['DIA']=EQTR2['DIA'].astype('category') print("Diameter of craters in equatorial region grouped") FREQ2=EQTR2['DIA'].value_counts(sort=True) print(FREQ2) PER2=EQTR2['DIA'].value_counts(sort=True, normalize=True)*100 print(PER2) #Data management of depth variable along with frequency and precentages EQTR2['DPTH']= pandas.cut(EQTR2.DEPTH_RIMFLOOR_TOPOG,[0.1,0.2,0.4,0.6,0.8,4.95]) EQTR2['DPTH']=EQTR2['DPTH'].astype('category') print("Depth of craters in equatorial region grouped") FREQ3=EQTR2['DPTH'].value_counts(sort=True) print(FREQ3) PER3=EQTR2['DPTH'].value_counts(sort=True, normalize=True)*100 print(PER3) #Summary statisstics of variables print("SUMMARY STATISTICS OF VARIABLES") print("Diameter of craters at equatorial region") stat1=EQTR2['DIAM_CIRCLE_IMAGE'].describe() print(stat1) print("median") mdn1=EQTR2['DIAM_CIRCLE_IMAGE'].median() print(mdn1) print("mode") md1=EQTR2['DIAM_CIRCLE_IMAGE'].mode() print(md1) print("Depths of craters in the equatorial region") stat2=EQTR2['DEPTH_RIMFLOOR_TOPOG'].describe() print(stat2) print("median") mdn2=EQTR2['DEPTH_RIMFLOOR_TOPOG'].median() print(mdn2) print("mode") md2=EQTR2['DEPTH_RIMFLOOR_TOPOG'].mode() print(md2) print("Number of ejecta layers on craters in the eqiatorial region") stat3=EQTR2['NUMBER_LAYERS'].describe() print(stat3) print("median") mdn3=EQTR2['NUMBER_LAYERS'].median() print(mdn3) print("mode") md3=EQTR2['NUMBER_LAYERS'].mode() print(md3) #Univariate graphs EQTR2['NUMBER_LAYERS']=EQTR2['NUMBER_LAYERS'].astype('category') seaborn.countplot(x="NUMBER_LAYERS", data=EQTR2) plt.xlabel('Number of ejecta layers') plt.title('Number of ejecta layers on craters in the Equatorial region of Mars surface') plt.show() seaborn.distplot(EQTR2['DIAM_CIRCLE_IMAGE'].dropna(), kde=False) plt.xlabel('Diameter of the craters in kms') plt.title('Distribution plot of the diamaters of the craters in the equtorial region') plt.show() seaborn.countplot(x="DIA", data=EQTR2) plt.xlabel('Diameter of the craters in kms') plt.title('Histogram of the diamaters of the craters grouped in the equtorial region') plt.show() seaborn.distplot(EQTR2['DEPTH_RIMFLOOR_TOPOG'].dropna(), kde=False) plt.xlabel('Depths of the craters in kms') plt.title('Distribution plot of the depths of the craters in the equtorial region') plt.show() seaborn.countplot(x="DPTH", data=EQTR2) plt.xlabel('Depths of the craters in kms') plt.title('Histogram of the depths of the craters grouped in the equtorial region') plt.show() #Bivariate graphs scat1=seaborn.regplot(x="LATITUDE_CIRCLE_IMAGE", y="DIAM_CIRCLE_IMAGE", fit_reg=False, data=EQTR2) plt.xlabel('Latitude') plt.ylabel('Diameter of craters in equatorial region') plt.title('scatter Plot: Latitude vs Diameters of craters') plt.show() scat2=seaborn.regplot(x="LATITUDE_CIRCLE_IMAGE", y="DEPTH_RIMFLOOR_TOPOG", fit_reg=False, data=EQTR2) plt.xlabel('Latitude') plt.ylabel('Depth of craters in equatorial region') plt.title('scatter Plot: Latitude vs Depth of craters') plt.show() seaborn.factorplot(x="EQR", y="DIAM_CIRCLE_IMAGE", data=EQTR2, kind="bar",ci=None) plt.xlabel('Latitude') plt.ylabel('Diameter of craters in equatorial region') plt.title('Bar Plot: Latitude vs Diameters of craters') plt.show() seaborn.factorplot(x="EQR", y="DEPTH_RIMFLOOR_TOPOG", data=EQTR2, kind="bar",ci=None) plt.xlabel('Latitude') plt.ylabel('Depth of craters in equatorial region') plt.title('Bar Plot: Latitude vs Depth of craters') plt.show()
Output:








The variables are grouped for better data management and plotting purposes. The summary statistics of the three variables of interest are found. It can be seen that the mean diameter of the craters at the equatorial region is 3.35 kms. This is in agreement with the hypothesis proposed at the beginning of the course that is the diameter of craters at the equatorial region are all small and is less than 8 kms. Similarly the depth (elevation) of the craters are also a low value that is 0.45 km. This can be connected to fact that most of the craters are having only 1 layer of ejecta.
It can be clearly seen from the histograms that all variables have right skewed distribution with the depth variable having a bi-modal right skewed nature. The variables are grouped into categories to find this.
Scatter plots are created with diameter and depth of craters as the response variables(y) and latitude as explanatory variables(x). While examining the scatter plot a relationship cannot be determined between the variable. So the variables are grouped and it can be concluded that the Diameter and latitude have a zigzag relationship. At the equatorial region its showing a bi-modal trend. The depth and latitude have a negative linear relationship in this region of mars.
1 note
·
View note
Text
Assignment, Week4
# Code:
import pandas import numpy import seaborn import matplotlib.pyplot as plt nesarc = pandas.read_csv ('nesarc_pds.csv' , low_memory=False) # load NESARC dataset
#Set PANDAS to show all columns in DataFrame pandas.set_option('display.max_columns', None) #Set PANDAS to show all rows in DataFrame pandas.set_option('display.max_rows', None)
nesarc.columns = map(str.upper , nesarc.columns)
pandas.set_option('display.float_format' , lambda x:'%f'%x)
# Change my variables to numeric nesarc['AGE'] = nesarc['AGE'].convert_objects(convert_numeric=True) nesarc['S3BQ4'] = nesarc['S3BQ4'].convert_objects(convert_numeric=True) nesarc['S3BQ1A5'] = nesarc['S3BQ1A5'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2B'] = nesarc['S3BD5Q2B'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2E'] = nesarc['S3BD5Q2E'].convert_objects(convert_numeric=True) nesarc['MAJORDEP12'] = nesarc['MAJORDEP12'].convert_objects(convert_numeric=True) nesarc['GENAXDX12'] = nesarc['GENAXDX12'].convert_objects(convert_numeric=True)
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BD5Q2B']==3) & (nesarc['S3BQ1A5']==1)] # Cannabis users both last 12 months and prior, ages 18-30 subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==2)] # Non-users, ages 18-30 subsetc2 = subset2.copy()
subset3 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30 subsetc3 = subset3.copy()
subset5 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==1)] # Cannabis users, ages 18-30 subsetc5 = subset5.copy()
# Frequency distributions of variables (groupby function) of the entire sample
print("Counts for cannabis use, variable S3BQ1A5 - Section 3B") cu1 = nesarc.groupby('S3BQ1A5').size() # Cannabis use counts print(cu1) print("Percentages for cannabis use, variable S3BQ1A5 - Section 3B") cu2 = nesarc.groupby('S3BQ1A5').size() * 100 / len(nesarc) # Cannabis use percentages print(cu2)
print("Counts for used cannabis in the last 12 months/prior to last 12 months/both time periods, variable S3BD5Q2B - Section 3B") uc1 = nesarc.groupby('S3BD5Q2B').size() # Used cannabis time periods counts print(uc1) print("Percentages for used cannabis in the last 12 months/prior to last 12 months/both time periods, S3BD5Q2B - Section 3B") uc2 = nesarc.groupby('S3BD5Q2B').size() * 100 / len(nesarc) # Used cannabis time periods percentages print(uc2)
nesarc.loc[(nesarc['S3BQ1A5']!=1) & (nesarc['S3BD5Q2E'].isnull()), 'S3BD5Q2E'] = 11
print("Counts for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc1 = nesarc.groupby('S3BD5Q2E').size() # Frequency of used cannabis counts print(fuc1) print("Percentages for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc2 = nesarc.groupby('S3BD5Q2E').size() * 100 / len(nesarc) # Frequency of used cannabis percentages print(fuc2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md1 = nesarc.groupby('MAJORDEP12').size() # Major depression diagnoses counts print(md1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md2 = nesarc.groupby('MAJORDEP12').size() * 100 / len(nesarc) # Major depression diagnoses percentages print(md2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga1 = nesarc.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts print(ga1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga2 = nesarc.groupby('GENAXDX12').size() * 100 / len(nesarc) # Generalized anxiety diagnoses percentages print(ga2)
# Frequency distributions of major depression and general anxiety diagnoses variables for both last 12 months and prior cannabis users, ages 18-30 (subset1)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu1 = subsetc1.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset1) print(mdu1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu2 = subsetc1.groupby('MAJORDEP12').size() * 100 / len(subsetc1) # Major depression diagnoses percentages (subset1) print(mdu2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau1 = subsetc1.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset1) print(gau1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau2 = subsetc1.groupby('GENAXDX12').size() * 100 / len(subsetc1) # Generalized anxiety diagnoses percentages (subset1) print(gau2)
# Frequency distributions of major depression and general anxiety diagnoses variables of non-users, ages 18-30 (subset2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn1 = subsetc2.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset2) print(mdn1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn2 = subsetc2.groupby('MAJORDEP12').size() * 100 / len(subsetc2) # Major depression diagnoses percentages (subset2) print(mdn2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan1 = subsetc2.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset2) print(gan1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan2 = subsetc2.groupby('GENAXDX12').size() * 100 / len(subsetc2) # Generalized anxiety diagnoses percentages (subset2) print(gan2)
###################################################################################################################################
# Quartile age split, cut function, 4 groups (18-21, 21-24, 24-27, 27-30)
subsetc3['AGE4GROUPS'] = pandas.qcut(subsetc3.AGE, 4, labels=["1=17-21","2=21-24","3=24-27","4=27-30"])
print("Counts for age splitted in 4 groups: 18-21, 21-24, 24-27, 27-30") age4g1 = subsetc3.groupby('AGE4GROUPS').size() print(age4g1)
print("Percentages for age splitted in 4 groups: 18-21, 21-24, 24-27, 27-30") age4g2 = subsetc3.groupby('AGE4GROUPS').size() * 100 / len(subsetc3) print(age4g2)
print("Counts of observations within each of the age group four categories") subsetc3['AGE4GROUPS'] = pandas.cut(subsetc3.AGE, [17, 21, 24, 27, 30]) print (pandas.crosstab(subsetc3['AGE4GROUPS'], subsetc3['AGE']))
# Frequency distribution of cannabis use variable for ages 18-30 (subset3) with 9 set to NaN, number of missing data
subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].replace(99, numpy.nan)
recode = {1: 1, 2: 2, 9: "NaN"} subsetc3['CUMD'] = subsetc3['S3BQ1A5'].map(recode)
print("Counts for cannabis use ages 18-30 with missing data set to NaN, variable S3BQ1A5 - Section 3B") cuy1 = subsetc3.groupby('CUMD').size() # Cannabis use counts (subset3) print(cuy1) print("Percentages for cannabis use age 18-30 with missing data set to NaN, variable S3BQ1A5 - Section 3B") cuy2 = subsetc3.groupby('CUMD').size() * 100 / len(subsetc3) # Cannabis use percentages (subset3) print(cuy2)
# Frequency distribution of monthly average cannabis used (when using the most) variable for ages 18-30 (subset5)
recode1 = {1: 10, 2: 9, 3: 8, 4: 7, 5: 6, 6: 5, 7: 4, 8: 3, 9: 2, 10: 1} # Dictionary with details of frequency variable reverse-recode subsetc5['CUFREQ'] = subsetc5['S3BD5Q2E'].map(recode1) # Change variable name from S3BD5Q2E to CUFREQ recode2 = {1: 30, 2: 25, 3: 14, 4: 6, 5: 3, 6: 1, 7: 0.8, 8: 0.5, 9: 0.3, 10: 0.1} # Monthly average cannabis used subsetc5['CUFREQMO'] = subsetc5['S3BD5Q2E'].map(recode2) # Change variable name from S3BD5Q2E to CUFREQMO
print("Counts for average cannabis used per month when using the most, variable CUFREQMO") fucy1 = subsetc5.groupby('CUFREQMO').size() # Frequency of used cannabis counts (subset5) print(fucy1) print("Percentages for average cannabis used per month when using the most, variable CUFREQMO") fucy2 = subsetc5.groupby('CUFREQMO').size() * 100 / len(subsetc5) # Frequency of used cannabis percentages (subset5) print(fucy2)
# Secondary variable creation, NUMJOPMOTH_EST, number of joints per month
subsetc5['NUMJOPMOTH_EST'] = subsetc5['CUFREQMO'] * subsetc5['S3BQ4'] subsetc4 = subsetc5[['IDNUM' , 'S3BQ4' , 'CUFREQMO' , 'NUMJOPMOTH_EST']] head30 = subsetc4.head(30) print("Number of cannabis joints smoked per month when using the most, first 30 observations, new variable NUMJOPMOTH_EST") print(head30)
# Frequency distribution for both major depression and general anxiety diagnoses, new variable cration NUMMDGENANX
subsetc1['NUMMDGENANX'] = subsetc1['MAJORDEP12'] + subsetc1['GENAXDX12']
print("Counts for major depression and general anxiety diagnoses in cannabis users, ages 18-30, variable NUMMDGENANX") ndg1 = subsetc1.groupby('NUMMDGENANX').size() print(ndg1) print("Percentages for major depression and general anxiety diagnoses in cannabis users, ages 18-30, variable NUMMDGENANX") ndg2 = subsetc1.groupby('NUMMDGENANX').size() * 100 / len(subsetc1) print(ndg2)
# Newly managed depression and anxiety variables, in cannabis users ages 18-30, both last 12 months and prior (subset1), define function
def DEPRESSIONANXIETY (row): if row['NUMMDGENANX'] == 0 : return 0 if row['NUMMDGENANX'] > 1 : return 1 if row['MAJORDEP12'] == 1 : return 2 if row['GENAXDX12'] == 1 : return 3
subsetc1['DEPRESSIONANXIETY'] = subsetc1.apply (lambda row: DEPRESSIONANXIETY (row), axis=1) subsetc6 = subsetc1[['IDNUM' , 'MAJORDEP12' , 'GENAXDX12' , 'NUMMDGENANX' , 'DEPRESSIONANXIETY']].copy() first30 = subsetc6.head(30) print("Depression and anxiety diagnoses counts for cannabis users in last 12 months and prior, ages 18-30, new variable DEPRESSIONANXIETY") print(first30)
####################################################################################################################################
print("Counts for average number of joints smoked per month, ages 18-30, variable NUMJOPMOTH_EST") njpm1 = subsetc5.groupby('NUMJOPMOTH_EST').size() print(njpm1) print("Percentages for average number of joints smoked per month, ages 18-30, variable NUMJOPMOTH_EST") njpm2 = subsetc5.groupby('NUMJOPMOTH_EST').size() * 100 / len(subsetc5) print(njpm2)
# Change format of S3BQ1A5, S3BD5Q2E, MAJORDEP12, GENAXDX12 to categorical
subsetc3['S3BQ1A5'] = subsetc3['S3BQ1A5'].astype('category') subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].astype('category') subsetc1['MAJORDEP12'] = subsetc1['MAJORDEP12'].astype('category') subsetc1['GENAXDX12'] = subsetc1['GENAXDX12'].astype('category')
# Change the numbers with strings, rename x-axis categories
subsetc1['MAJORDEP12'] = subsetc1['MAJORDEP12'].cat.rename_categories(["No","Yes"]) subsetc1['GENAXDX12'] = subsetc1['GENAXDX12'].cat.rename_categories(["No","Yes"]) subsetc3['S3BQ1A5'] = subsetc3['S3BQ1A5'].cat.rename_categories(["Yes","No","Unknown"]) subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].cat.rename_categories(["Every day","Nearly every day","3-4 times/week","1-2 times/week","2-3 times/month","Once a month","7-11 times/year","3-6 times/year","2 times/year","Once a year"])
# Univariate bar chart for categorical variables, S3BQ1A5, S3BD5Q2E, MAJORDEP12, GENAXDX12, which stand for cannabis use, frequency of use, major depression and general anxiety
seaborn.countplot(x='S3BQ1A5', data=subsetc3) plt.xlabel('Cannabis use in ages 18-30') plt.title('Cannabis use, ages 18-30') plt.show() print ('Describe cannabis use variable') djpm2 = subsetc3['S3BQ1A5'].describe() print (djpm2)
plt.figure(figsize=(12,4)) # Change plot size ax5 = seaborn.countplot(x='S3BD5Q2E', data=subsetc5) ax5.set_xticklabels(ax5.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use in ages 18-30') plt.title('Frequency of cannabis use, ages 18-30') plt.show() print ('Describe frequency of cannabis use variable') djpm5 = subsetc5['S3BD5Q2E'].describe() print (djpm5)
seaborn.countplot(x='MAJORDEP12', data=subsetc1) plt.xlabel('Major depression diagnoses in last 12 months') plt.title('Major depression diagnosed in cannabis users (both last 12 months and prior), ages 18-30') plt.show() print ('Describe major depression variable') djpm3 = subsetc1['MAJORDEP12'].describe() print (djpm3) seaborn.countplot(x='GENAXDX12', data=subsetc1) plt.xlabel('General anxiety diagnoses in last 12 months') plt.title('General anxiety diagnosed in cannabis users (both last 12 months and prior), ages 18-30') plt.show() print ('Describe general anxiety variable') djpm4 = subsetc1['GENAXDX12'].describe() print (djpm4)
# Univariate bar chart for quantitative variable, NUMJOPMOTH_EST, that stands for the number of joints smoked per month when using the most, ages 18-30
plt.figure(figsize=(10,4)) # Change plot size seaborn.distplot(subsetc5["NUMJOPMOTH_EST"].dropna(), kde=False) plt.xlabel('Number of joints smoked per month') plt.title('Estimated number of joints smoked per month by cannabis users, ages 18-30') plt.show()
# Center and spread measurements
print ('Spread') std1 = subsetc5['NUMJOPMOTH_EST'].std() print (std1)
print ('Mode') mode1 = subsetc5['NUMJOPMOTH_EST'].mode() print (mode1)
print ('Mean') mean1 = subsetc5['NUMJOPMOTH_EST'].mean() print (mean1)
print ('Median') median1 = subsetc5['NUMJOPMOTH_EST'].median() print (median1)
# Plot for groups of total number of joints smoked per month, variable NUMJOPMOTH_EST
subsetc5['NUMJOPMOTH_EST'] = pandas.cut(subsetc5.NUMJOPMOTH_EST, [0, 1, 10, 20, 30, 50, 70, 90, 110, 130, 150, 200, 250, 300, 2970]) # Split the number into groups subsetc5['NUMJOPMOTH_EST'] = subsetc5['NUMJOPMOTH_EST'].astype('category') # Rename x-axis categories of the plot subsetc5['NUMJOPMOTH_EST'] = subsetc5['NUMJOPMOTH_EST'].cat.rename_categories(["<1","1-10","11-20","21-30","31-50","51-70","71-90","90-110","111-130","131-150","151-200","201-250","251-300",">300"]) plt.figure(figsize=(10,4)) # Change plot size ax = seaborn.countplot(x='NUMJOPMOTH_EST', data=subsetc5) ax.set_xticklabels(ax.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Number of joints smoked per month') plt.title('Estimated number of joints smoked per month by cannabis users, ages 18-30') plt.show()
print ('Describe number of joints smoked per month') djpm1 = subsetc5['NUMJOPMOTH_EST'].describe() print (djpm1)
# Bivariate bar graph C->Q, major depression as response variable and number of joints smoked per month as explanatory variable
nesarc['MAJORDEP12'] = nesarc['MAJORDEP12'].convert_objects(convert_numeric=True)
plt.figure(figsize=(10,4)) # Change plot size ax1 = seaborn.factorplot(x="NUMJOPMOTH_EST", y="MAJORDEP12", data=subsetc5, kind="bar", ci=None) ax1.set_xticklabels(ax.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Joints smoked per Month') plt.ylabel('Proportion of major depression') plt.show()
# Bivariate bar graph C->Q, general anxiety as response variable and number of joints smoked per month as explanatory variable
nesarc['GENAXDX12'] = nesarc['GENAXDX12'].convert_objects(convert_numeric=True)
plt.figure(figsize=(10,4)) # Change plot size ax2 = seaborn.factorplot(x="NUMJOPMOTH_EST", y="GENAXDX12", data=subsetc5, kind="bar", ci=None) ax2.set_xticklabels(ax.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Joints smoked per Month') plt.ylabel('Proportion of general anxiety') plt.show()
nesarc['S3BQ1A5'] = nesarc['S3BQ1A5'].convert_objects(convert_numeric=True) subsetc3['S3BQ1A5']=subsetc3['S3BQ1A5'].replace(9, numpy.nan)
# Bivariate bar graph C->C, major depression as response variable and cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(10,4)) # Change plot size seaborn.factorplot(x="S3BQ1A5", y="MAJORDEP12", data=subsetc3, kind="bar", ci=None) plt.xlabel('Cannabis use ages 18-30') plt.ylabel('Proportion of major depression') plt.show()
# Bivariate bar graph C->C, general anxiety as response variable and cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(10,4)) # Change plot size seaborn.factorplot(x="S3BQ1A5", y="GENAXDX12", data=subsetc3, kind="bar", ci=None) plt.xlabel('Cannabis use ages 18-30') plt.ylabel('Proportion of general anxiety') plt.show()
# Change frequency variable type to categorical
subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].astype('category')
# Bivariate bar graph C->C, major depression as response variable and frequency of cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(12,4)) # Change plot size ax3 = seaborn.factorplot(x="S3BD5Q2E", y="MAJORDEP12", data=subsetc5, kind="bar", ci=None) ax3.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use ages 18-30') plt.ylabel('Proportion of major depression') plt.show()
# Bivariate bar graph C->C, general anxiety as response variable and frequency of cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(12,4)) # Change plot size ax4 = seaborn.factorplot(x="S3BD5Q2E", y="GENAXDX12", data=subsetc5, kind="bar", ci=None) ax4.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use ages 18-30') plt.ylabel('Proportion of general anxiety') plt.show()
# Output Discussion with Github Links for output images
# Visualizing Data
## Preview In the final assignment are presented visualized data, taken from NESARC codebook, in order to examine the correlation between cannabis use and mental disorders such as major depression and general anxiety diagnosed in the last 12 months in a sample of 9535 U.S. young adults, aged from 18 to 30 years old. I used Spyder IDE to create both univariate and bivariate bar charts for the selected variables. More specifically, with variable ‘AGE’ between 18 and 30, I built unvariate graphs for categorical variables **‘S3BQ1A5’** which represents cannabis use, **‘S3BD5Q2E’** which is frequency of this use, **‘MAJORDEP12’** that stands for major depression diagnosis in the last 12 months and **‘GENAXDX12’** that indicates general anxiety diagnosis in the same period. In addition, you will find another univariate graph for the quantitative variable **‘NUMJOPMOTH_EST’**, which I created in my previous assignment by multiplying frequency of cannabis use and average quantity of joints smoked, in order to estimate the total number of joints smoked per month by the individuals. As far as the bivariate graphs are concerned, I chose to examine visualized the association between cannabis use (C->C) and both mentioned disorders and additionally the relationship between frequency (C->C) and quantity (Q->C) of this use with both depression and anxiety. Thus, bar charts were created combining variables **‘S3BQ1A5’** (cannabis use), ‘S3BD5Q2E’ (frequency of use) and **‘NUMJOPMOTH_EST’** (quantity of joints) with variables **‘MAJORDEP12’** (major depression) and **‘GENAXDX12’** (general anxiety). Concluding, for the quantitative variable both center and spread were measured and describe function was used in order to examine useful information, about the selected categorical variables.
## Output
### Univariate graphs: 
A random sample of 9535 U.S. young adults, aged 18-30, were asked, as a part of NESARC survey, the following question: “Have you ever used cannabis?” A percentage of 25.29% (or 7042 individuals) answered “Yes”, whereas 73.85% (or about 2500 individuals) answered “No” which was the most frequent answer. Also a significantly small percentage of 0.84%, fell into category 9 (“Unknown“) which is our missing data.

To the question of “How often did you use cannabis when using the most?”, the top answer was “Every day”, since 534 individuals fell into this category, followed by “Once a year” category with approximately 400 individuals. Less than 100 people chose “7-11 times per year” category, which was the least frequent answer.

Of the total number of participants (18-30) who answered “Yes” to the question of cannabis use, only those who were smoking marijuana in last 12 months and prior were taken into consideration for the next two questions.
To the question of “Have you been diagnosed with non-hierarchical major depression in the last 12 months?”, about 660 participants or 79.04% answered “No” which was the most frequent answer, whereas 175 or 20.95% fell into “Yes”.
For the question, ”Have you been diagnosed with non-hierarchical generalized anxiety in the last 12 months?”, 802 individuals or 96.04% answered “No“ that was our top answer, while only 33 or 3.95% chose “Yes“.

For the estimated number of joints smoked per month by cannabis users, ages 18-30, it noticeable from the graph that there was a skewed-right distribution. The spread or the standard deviation of the variable is extremely large which indicates a large variety of answers among the participants. The three main numerical measures of the center of the distribution are the mode, the median, and the mean. Here we can see that mode is equal to 0.1 and it was the most common occurring value in the distribution, which means that most of participants smoked less than 1 joint per month. The mean is equal to 70.1 which indicates that cannabis users smoked about 70 joints per month on average and the median or the middle value is 6.

Estimated number of joints smoked per month binned to groups as illustrated above. Another way of visualizing the distribution of variable **‘NUMJOPMOTH_EST’**. We can see that most individuals, about 990, smoked less than one joint per month and the shape of the distribution is right-skewed.
### Bivariate graphs:  
In the bar charts above we can see the relationship between quantity of joints smoked per month by cannabis user, aged 18 to 30 years old, and both major depression (first) and general anxiety (second) diagnoses in the last 12 months (Q->C). The explanatory variable is quantity of joints (quantitative), while the response variables are depression and anxiety diagnoses (categorical). There is a slightly increasing trend in the first graph, but not in the second.
 
In the graphs presented above we can see the correlation between frequency of cannabis use and both major depression and general anxiety (C->C). The explanatory variable is frequency of cannabis use (categorical), while the response variables are depression and anxiety diagnoses (categorical). Again, for the first graph we have a right-skewed distribution, which indicates that the more an individual smoked cannabis, the better were the chances to get diagnosed with depression. However, we cannot support the same as far as anxiety is concerned, which appears to have a more raffle and abnormal distribution.
 
The graphs presented above illustrate the association between cannabis use and both major depression and general anxiety diagnoses in young adults, aged from 18 to 30 years old, in the last 12 months (C->C). The explanatory variable is cannabis use (categorical) and the response variables are depression and anxiety diagnoses (categorical).
## Summary To sum up, looking through the the last graphs, it can be noticed that there are some slight differences between the percentages of cannabis users compared to non-users. Major depression cases in cannabis users young adults (20.95%) seem to be slightly more than double compared to those of non-users (8.42%).In addition, general anxiety diagnoses in cannabis users (3.95%) appear to be also marginally more than double in comparison to the non-users (1.63%). It could be supported that there is a relative association between cannabis and such mental disorders, thus cannabis use increases the likelihood of meeting criteria for depression or general anxiety in the future. However, the sample is extremely small and it is unclear how representative it is, making the findings less reliable, since a large amount of error may be involved.
0 notes
Text
Week 3 Submission
Making Data Management Decisions
Preview In this assignment you will find three data managed variables, provided from NESARC codebook, as frequency distributions. For the variable ‘AGE’, I decided to split the ages of the observations in four categories (18-21, 21-24, 24-27, 27-30), in order to examine the age frequency distribution of the sample into quartiles. Furthermore, I included missing data in the cannabis use variable distribution for ages 18 to 30, with the row ‘Unknown’ set to ‘nan’. In addition, after recoding variable ‘S3BD5Q2E’, which represents average rate of cannabis use, I created a new variable called ‘CUFREQMO’ that indicates how many days an individual used cannabis per month, when using the most. Moreover, once I ran frequency distribution for the variable ‘CUFREQMO’ and calculated the counts and the percentages of this use per month, I created a secondary variable with the name ‘NUMJOPMOTH_EST’. This variable estimates the quantity of cannabis joints smoked per month for the first 30 participants (18-30) when using the most, by multiplying the amount of joints smoked per day (variable ‘S3BQ4’) with the variable ‘CUFREQMO’ , which indicates the number of days an individual smoked per month. I finished with the creation of one more variable called ‘NUMMDGENANX’ that stands for the total number of participants who were diagnosed with either major depression, general anxiety or both illnesses. After examining the frequency distribution of this variable, I used the define function to create one last variable with the name ‘DEPRESSIONANXIETY’, which characterizes the mental state of the first 30 random cannabis users aged 18 to 30, as far as these two disorders are concerned. For the code and the output i used Jupyter Notebook.
Firstly, a new variable called ‘AGE4GROUPS’ was necessary to be created in order to divide participants, aged between 18 and 30 years old, into four age groups: 18-21, 21-24, 24-27 and 27-30. From the frequency distribution it can be noticed that about 30.73% were between 18 and 21 years old, 23.78% were between 21 and 24, 20.65% fell into 24-27 group and 24.82% fell into 27-30 group.
For the frequency distribution of variable ‘S3BQ1A5′ the missing data is included and option 9 is set to ‘NaN’, which represents the choice ‘Unknown’. As we can see the number of the missing data is 81 individuals and the proportion is 0.84% which is significantly small. Regarding the new variable with the name ’CUFREQMO’ which I created, the distribution results show that about 15.96% smoked cannabis less than once per year, whereas approximately 22.13% used to smoke cannabis in an everyday basis. The rest of the cannabis use rates vary from about 2.65% for less than once per month, to 11.6% for an average use of 6 times per month.
Taking into account the new variable ‘NUMJOPMOTH_EST’ which I created, the output illustrates the estimation of the quantity of joints smoked by young adults aged 18 to 30 per month, by multiplying the quantity of joints smoked per day with the total number of days that they used cannabis per month. By examining the first 30 observations of the sample, it is noticeable that the majority of individuals smoked approximately less than 10 joints per month. On the other hand there were also some participants who were smoking more than 100 joints monthly, since individuals with ID numbers 511 and 506 for example, peaked at 180 and 150 joints per month respectively.
As far as the new secondary variable ‘NUMMDGENANX’ is concerned, it is obvious that around 77% of cannabis users aged between 18 and 30 were mentally healthy in the last 12 months, after using marijuana for more than 1 year, while a percentage of 21.07% were diagnosed with either major depression or general anxiety. Only 1.91% of them were diagnosed with both mental disorders mentioned above. Finally, regarding the first 30 observations table, presented in the end, the define function used, returns 0 if an individual had no problem with either depression or anxiety in the last 12 months, 1 if an individual met the criteria for both disorders, 2 if an individual was diagnosed only with major depression and 3 if an individual was diagnosed only with general anxiety. For the results, a new variable named ‘DEPRESSIONANXIETY’ was created.
Code-
import pandas import numpy import seaborn import matplotlib.pyplot as plt nesarc = pandas.read_csv ('nesarc_pds.csv' , low_memory=False) # load NESARC dataset
#Set PANDAS to show all columns in DataFrame pandas.set_option('display.max_columns', None) #Set PANDAS to show all rows in DataFrame pandas.set_option('display.max_rows', None)
nesarc.columns = map(str.upper , nesarc.columns)
pandas.set_option('display.float_format' , lambda x:'%f'%x)
# Change my variables to numeric nesarc['AGE'] = nesarc['AGE'].convert_objects(convert_numeric=True) nesarc['S3BQ4'] = nesarc['S3BQ4'].convert_objects(convert_numeric=True) nesarc['S3BQ1A5'] = nesarc['S3BQ1A5'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2B'] = nesarc['S3BD5Q2B'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2E'] = nesarc['S3BD5Q2E'].convert_objects(convert_numeric=True) nesarc['MAJORDEP12'] = nesarc['MAJORDEP12'].convert_objects(convert_numeric=True) nesarc['GENAXDX12'] = nesarc['GENAXDX12'].convert_objects(convert_numeric=True)
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BD5Q2B']==3) & (nesarc['S3BQ1A5']==1)] # Cannabis users both last 12 months and prior, ages 18-30 subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==2)] # Non-users, ages 18-30 subsetc2 = subset2.copy()
subset3 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30 subsetc3 = subset3.copy()
subset5 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==1)] # Cannabis users, ages 18-30 subsetc5 = subset5.copy()
# Frequency distributions of variables (groupby function) of the entire sample
print("Counts for cannabis use, variable S3BQ1A5 - Section 3B") cu1 = nesarc.groupby('S3BQ1A5').size() # Cannabis use counts print(cu1) print("Percentages for cannabis use, variable S3BQ1A5 - Section 3B") cu2 = nesarc.groupby('S3BQ1A5').size() * 100 / len(nesarc) # Cannabis use percentages print(cu2)
print("Counts for used cannabis in the last 12 months/prior to last 12 months/both time periods, variable S3BD5Q2B - Section 3B") uc1 = nesarc.groupby('S3BD5Q2B').size() # Used cannabis time periods counts print(uc1) print("Percentages for used cannabis in the last 12 months/prior to last 12 months/both time periods, S3BD5Q2B - Section 3B") uc2 = nesarc.groupby('S3BD5Q2B').size() * 100 / len(nesarc) # Used cannabis time periods percentages print(uc2)
nesarc.loc[(nesarc['S3BQ1A5']!=1) & (nesarc['S3BD5Q2E'].isnull()), 'S3BD5Q2E'] = 11
print("Counts for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc1 = nesarc.groupby('S3BD5Q2E').size() # Frequency of used cannabis counts print(fuc1) print("Percentages for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc2 = nesarc.groupby('S3BD5Q2E').size() * 100 / len(nesarc) # Frequency of used cannabis percentages print(fuc2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md1 = nesarc.groupby('MAJORDEP12').size() # Major depression diagnoses counts print(md1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md2 = nesarc.groupby('MAJORDEP12').size() * 100 / len(nesarc) # Major depression diagnoses percentages print(md2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga1 = nesarc.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts print(ga1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga2 = nesarc.groupby('GENAXDX12').size() * 100 / len(nesarc) # Generalized anxiety diagnoses percentages print(ga2)
# Frequency distributions of major depression and general anxiety diagnoses variables for both last 12 months and prior cannabis users, ages 18-30 (subset1)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu1 = subsetc1.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset1) print(mdu1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu2 = subsetc1.groupby('MAJORDEP12').size() * 100 / len(subsetc1) # Major depression diagnoses percentages (subset1) print(mdu2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau1 = subsetc1.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset1) print(gau1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau2 = subsetc1.groupby('GENAXDX12').size() * 100 / len(subsetc1) # Generalized anxiety diagnoses percentages (subset1) print(gau2)
# Frequency distributions of major depression and general anxiety diagnoses variables of non-users, ages 18-30 (subset2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn1 = subsetc2.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset2) print(mdn1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn2 = subsetc2.groupby('MAJORDEP12').size() * 100 / len(subsetc2) # Major depression diagnoses percentages (subset2) print(mdn2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan1 = subsetc2.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset2) print(gan1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan2 = subsetc2.groupby('GENAXDX12').size() * 100 / len(subsetc2) # Generalized anxiety diagnoses percentages (subset2) print(gan2)
###################################################################################################################################
# Quartile age split, cut function, 4 groups (18-21, 21-24, 24-27, 27-30)
subsetc3['AGE4GROUPS'] = pandas.qcut(subsetc3.AGE, 4, labels=["1=17-21","2=21-24","3=24-27","4=27-30"])
print("Counts for age splitted in 4 groups: 18-21, 21-24, 24-27, 27-30") age4g1 = subsetc3.groupby('AGE4GROUPS').size() print(age4g1)
print("Percentages for age splitted in 4 groups: 18-21, 21-24, 24-27, 27-30") age4g2 = subsetc3.groupby('AGE4GROUPS').size() * 100 / len(subsetc3) print(age4g2)
print("Counts of observations within each of the age group four categories") subsetc3['AGE4GROUPS'] = pandas.cut(subsetc3.AGE, [17, 21, 24, 27, 30]) print (pandas.crosstab(subsetc3['AGE4GROUPS'], subsetc3['AGE']))
# Frequency distribution of cannabis use variable for ages 18-30 (subset3) with 9 set to NaN, number of missing data
subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].replace(99, numpy.nan)
recode = {1: 1, 2: 2, 9: "NaN"} subsetc3['CUMD'] = subsetc3['S3BQ1A5'].map(recode)
print("Counts for cannabis use ages 18-30 with missing data set to NaN, variable S3BQ1A5 - Section 3B") cuy1 = subsetc3.groupby('CUMD').size() # Cannabis use counts (subset3) print(cuy1) print("Percentages for cannabis use age 18-30 with missing data set to NaN, variable S3BQ1A5 - Section 3B") cuy2 = subsetc3.groupby('CUMD').size() * 100 / len(subsetc3) # Cannabis use percentages (subset3) print(cuy2)
# Frequency distribution of monthly average cannabis used (when using the most) variable for ages 18-30 (subset5)
recode1 = {1: 10, 2: 9, 3: 8, 4: 7, 5: 6, 6: 5, 7: 4, 8: 3, 9: 2, 10: 1} # Dictionary with details of frequency variable reverse-recode subsetc5['CUFREQ'] = subsetc5['S3BD5Q2E'].map(recode1) # Change variable name from S3BD5Q2E to CUFREQ recode2 = {1: 30, 2: 25, 3: 14, 4: 6, 5: 3, 6: 1, 7: 0.8, 8: 0.5, 9: 0.3, 10: 0.1} # Monthly average cannabis used subsetc5['CUFREQMO'] = subsetc5['S3BD5Q2E'].map(recode2) # Change variable name from S3BD5Q2E to CUFREQMO
print("Counts for average cannabis used per month when using the most, variable CUFREQMO") fucy1 = subsetc5.groupby('CUFREQMO').size() # Frequency of used cannabis counts (subset5) print(fucy1) print("Percentages for average cannabis used per month when using the most, variable CUFREQMO") fucy2 = subsetc5.groupby('CUFREQMO').size() * 100 / len(subsetc5) # Frequency of used cannabis percentages (subset5) print(fucy2)
# Secondary variable creation, NUMJOPMOTH_EST, number of joints per month
subsetc5['NUMJOPMOTH_EST'] = subsetc5['CUFREQMO'] * subsetc5['S3BQ4'] subsetc4 = subsetc5[['IDNUM' , 'S3BQ4' , 'CUFREQMO' , 'NUMJOPMOTH_EST']] head30 = subsetc4.head(30) print("Number of cannabis joints smoked per month when using the most, first 30 observations, new variable NUMJOPMOTH_EST") print(head30)
# Frequency distribution for both major depression and general anxiety diagnoses, new variable cration NUMMDGENANX
subsetc1['NUMMDGENANX'] = subsetc1['MAJORDEP12'] + subsetc1['GENAXDX12']
print("Counts for major depression and general anxiety diagnoses in cannabis users, ages 18-30, variable NUMMDGENANX") ndg1 = subsetc1.groupby('NUMMDGENANX').size() print(ndg1) print("Percentages for major depression and general anxiety diagnoses in cannabis users, ages 18-30, variable NUMMDGENANX") ndg2 = subsetc1.groupby('NUMMDGENANX').size() * 100 / len(subsetc1) print(ndg2)
# Newly managed depression and anxiety variables, in cannabis users ages 18-30, both last 12 months and prior (subset1), define function
def DEPRESSIONANXIETY (row): if row['NUMMDGENANX'] == 0 : return 0 if row['NUMMDGENANX'] > 1 : return 1 if row['MAJORDEP12'] == 1 : return 2 if row['GENAXDX12'] == 1 : return 3
subsetc1['DEPRESSIONANXIETY'] = subsetc1.apply (lambda row: DEPRESSIONANXIETY (row), axis=1) subsetc6 = subsetc1[['IDNUM' , 'MAJORDEP12' , 'GENAXDX12' , 'NUMMDGENANX' , 'DEPRESSIONANXIETY']].copy() first30 = subsetc6.head(30) print("Depression and anxiety diagnoses counts for cannabis users in last 12 months and prior, ages 18-30, new variable DEPRESSIONANXIETY") print(first30)
####################################################################################################################################
print("Counts for average number of joints smoked per month, ages 18-30, variable NUMJOPMOTH_EST") njpm1 = subsetc5.groupby('NUMJOPMOTH_EST').size() print(njpm1) print("Percentages for average number of joints smoked per month, ages 18-30, variable NUMJOPMOTH_EST") njpm2 = subsetc5.groupby('NUMJOPMOTH_EST').size() * 100 / len(subsetc5) print(njpm2)
# Change format of S3BQ1A5, S3BD5Q2E, MAJORDEP12, GENAXDX12 to categorical
subsetc3['S3BQ1A5'] = subsetc3['S3BQ1A5'].astype('category') subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].astype('category') subsetc1['MAJORDEP12'] = subsetc1['MAJORDEP12'].astype('category') subsetc1['GENAXDX12'] = subsetc1['GENAXDX12'].astype('category')
# Change the numbers with strings, rename x-axis categories
subsetc1['MAJORDEP12'] = subsetc1['MAJORDEP12'].cat.rename_categories(["No","Yes"]) subsetc1['GENAXDX12'] = subsetc1['GENAXDX12'].cat.rename_categories(["No","Yes"]) subsetc3['S3BQ1A5'] = subsetc3['S3BQ1A5'].cat.rename_categories(["Yes","No","Unknown"]) subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].cat.rename_categories(["Every day","Nearly every day","3-4 times/week","1-2 times/week","2-3 times/month","Once a month","7-11 times/year","3-6 times/year","2 times/year","Once a year"])
# Univariate bar chart for categorical variables, S3BQ1A5, S3BD5Q2E, MAJORDEP12, GENAXDX12, which stand for cannabis use, frequency of use, major depression and general anxiety
seaborn.countplot(x='S3BQ1A5', data=subsetc3) plt.xlabel('Cannabis use in ages 18-30') plt.title('Cannabis use, ages 18-30') plt.show() print ('Describe cannabis use variable') djpm2 = subsetc3['S3BQ1A5'].describe() print (djpm2)
plt.figure(figsize=(12,4)) # Change plot size ax5 = seaborn.countplot(x='S3BD5Q2E', data=subsetc5) ax5.set_xticklabels(ax5.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use in ages 18-30') plt.title('Frequency of cannabis use, ages 18-30') plt.show() print ('Describe frequency of cannabis use variable') djpm5 = subsetc5['S3BD5Q2E'].describe() print (djpm5)
seaborn.countplot(x='MAJORDEP12', data=subsetc1) plt.xlabel('Major depression diagnoses in last 12 months') plt.title('Major depression diagnosed in cannabis users (both last 12 months and prior), ages 18-30') plt.show() print ('Describe major depression variable') djpm3 = subsetc1['MAJORDEP12'].describe() print (djpm3) seaborn.countplot(x='GENAXDX12', data=subsetc1) plt.xlabel('General anxiety diagnoses in last 12 months') plt.title('General anxiety diagnosed in cannabis users (both last 12 months and prior), ages 18-30') plt.show() print ('Describe general anxiety variable') djpm4 = subsetc1['GENAXDX12'].describe() print (djpm4)
# Univariate bar chart for quantitative variable, NUMJOPMOTH_EST, that stands for the number of joints smoked per month when using the most, ages 18-30
plt.figure(figsize=(10,4)) # Change plot size seaborn.distplot(subsetc5["NUMJOPMOTH_EST"].dropna(), kde=False) plt.xlabel('Number of joints smoked per month') plt.title('Estimated number of joints smoked per month by cannabis users, ages 18-30') plt.show()
# Center and spread measurements
print ('Spread') std1 = subsetc5['NUMJOPMOTH_EST'].std() print (std1)
print ('Mode') mode1 = subsetc5['NUMJOPMOTH_EST'].mode() print (mode1)
print ('Mean') mean1 = subsetc5['NUMJOPMOTH_EST'].mean() print (mean1)
print ('Median') median1 = subsetc5['NUMJOPMOTH_EST'].median() print (median1)
# Plot for groups of total number of joints smoked per month, variable NUMJOPMOTH_EST
subsetc5['NUMJOPMOTH_EST'] = pandas.cut(subsetc5.NUMJOPMOTH_EST, [0, 1, 10, 20, 30, 50, 70, 90, 110, 130, 150, 200, 250, 300, 2970]) # Split the number into groups subsetc5['NUMJOPMOTH_EST'] = subsetc5['NUMJOPMOTH_EST'].astype('category') # Rename x-axis categories of the plot subsetc5['NUMJOPMOTH_EST'] = subsetc5['NUMJOPMOTH_EST'].cat.rename_categories(["<1","1-10","11-20","21-30","31-50","51-70","71-90","90-110","111-130","131-150","151-200","201-250","251-300",">300"]) plt.figure(figsize=(10,4)) # Change plot size ax = seaborn.countplot(x='NUMJOPMOTH_EST', data=subsetc5) ax.set_xticklabels(ax.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Number of joints smoked per month') plt.title('Estimated number of joints smoked per month by cannabis users, ages 18-30') plt.show()
print ('Describe number of joints smoked per month') djpm1 = subsetc5['NUMJOPMOTH_EST'].describe() print (djpm1)
Screenshots of Outputs-




0 notes
Text
Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Pubmed: http://dlvr.it/QnV0pN
0 notes
Text
Favorite tweets
伊藤万理華さんの写真集も発売され、YouTubeに展覧会での映像作品もアップされたこのタイミングに、改めてMdN1月号のこの付録ブックレット「伊藤万理華が乃木坂46に残したクリエイティブ」をお読み頂きたいです!在庫を急遽、Amazonに出しました! https://t.co/OarpNrEKMm
— 本信光理 (@mdn_hikari) February 27, 2018
from http://twitter.com/mdn_hikari via IFTTT
0 notes
Text
On Twitter ...
発売中のMdN1月号で伊藤万理華さんの連載「MARIKA meets CREATORS」の最後の対談に呼んでいただきました。「咄嗟」や「行くあて」など懐かしい話をたくさんしてます。付録ブックレットの方にもインタビューを掲載させていただいてますのでそちらもぜひご覧ください。 http://pic.twitter.com/RDYPu52Y17
— 湯浅弘章 (@HiroYuasa) December 7, 2017
from Twitter https://twitter.com/sore_miryoku_ December 07, 2017 at 08:10PM via IFTTT
0 notes
Text
【乃木坂46】伊藤万理華ロングインタビュー、『MdN1月号』に1万字で掲載予定!
var script = document.createElement(‘script’); script.src = “http://manga.boy.jp/blog.livedoor.com/1111_nogizaka46matome.dreamlog.jp.js”; document.getElementsByTagName(‘body’)[0].appendChild(script); 495 君の名は :2017/11/17(金) 20:33:09.57 ID:igDt61LM0.net うおおおとお!ロングインタビューくそ楽しみ 498 君の名は :2017/11/17(金) 21:43:47.84 ID:+yJ5d+8kp.net これかな 本信光理@mdn_hikari この前、伊藤万理華さんのロングインタビューをした。…
View On WordPress
0 notes
Text
【乃木坂46】『伊藤万理華が乃木坂46に残したクリエイティブ』次号『MdN1月号』の内容がアツい・・・
【乃木坂46】『伊藤万理華が乃木坂46に残したクリエイティブ』次号『MdN1月号』の内容がアツい・・・ – 乃木坂46まとめ 1/46 続きを読む Source: なんでも通信談
View On WordPress
0 notes
Photo

I know I can' sing cover art by #ChevyBoss "This mixtape original dropped on datpiff February early 2016 I had just dropped MDN1 and wanted to keep something out to share with people . The original cover was pink because of that reason kind of dull and when I wanted to do the rerelease I wanted something more bright and dark to which is why the burnt orange to dark red . It's kind of a psychedelic moment so the lettering is some kind of 80's style and so is the music it's more instrumental and a lot of sampling in the tape it's blurry vintage to me this was my second mixtape I had done it in a basement majority of it on the south side of chicago it was dope time" - ChevyBoss (at Far Southwest Side, Chicago, Illinois)
0 notes
Text
Assignment Week 3
## Code:
import pandas import numpy import seaborn import matplotlib.pyplot as plt nesarc = pandas.read_csv ('nesarc_pds.csv' , low_memory=False) # load NESARC dataset
#Set PANDAS to show all columns in DataFrame pandas.set_option('display.max_columns', None) #Set PANDAS to show all rows in DataFrame pandas.set_option('display.max_rows', None)
nesarc.columns = map(str.upper , nesarc.columns)
pandas.set_option('display.float_format' , lambda x:'%f'%x)
# Change my variables to numeric nesarc['AGE'] = nesarc['AGE'].convert_objects(convert_numeric=True) nesarc['S3BQ4'] = nesarc['S3BQ4'].convert_objects(convert_numeric=True) nesarc['S3BQ1A5'] = nesarc['S3BQ1A5'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2B'] = nesarc['S3BD5Q2B'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2E'] = nesarc['S3BD5Q2E'].convert_objects(convert_numeric=True) nesarc['MAJORDEP12'] = nesarc['MAJORDEP12'].convert_objects(convert_numeric=True) nesarc['GENAXDX12'] = nesarc['GENAXDX12'].convert_objects(convert_numeric=True)
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BD5Q2B']==3) & (nesarc['S3BQ1A5']==1)] # Cannabis users both last 12 months and prior, ages 18-30 subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==2)] # Non-users, ages 18-30 subsetc2 = subset2.copy()
subset3 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30 subsetc3 = subset3.copy()
subset5 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==1)] # Cannabis users, ages 18-30 subsetc5 = subset5.copy()
# Frequency distributions of variables (groupby function) of the entire sample
print("Counts for cannabis use, variable S3BQ1A5 - Section 3B") cu1 = nesarc.groupby('S3BQ1A5').size() # Cannabis use counts print(cu1) print("Percentages for cannabis use, variable S3BQ1A5 - Section 3B") cu2 = nesarc.groupby('S3BQ1A5').size() * 100 / len(nesarc) # Cannabis use percentages print(cu2)
print("Counts for used cannabis in the last 12 months/prior to last 12 months/both time periods, variable S3BD5Q2B - Section 3B") uc1 = nesarc.groupby('S3BD5Q2B').size() # Used cannabis time periods counts print(uc1) print("Percentages for used cannabis in the last 12 months/prior to last 12 months/both time periods, S3BD5Q2B - Section 3B") uc2 = nesarc.groupby('S3BD5Q2B').size() * 100 / len(nesarc) # Used cannabis time periods percentages print(uc2)
nesarc.loc[(nesarc['S3BQ1A5']!=1) & (nesarc['S3BD5Q2E'].isnull()), 'S3BD5Q2E'] = 11
print("Counts for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc1 = nesarc.groupby('S3BD5Q2E').size() # Frequency of used cannabis counts print(fuc1) print("Percentages for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc2 = nesarc.groupby('S3BD5Q2E').size() * 100 / len(nesarc) # Frequency of used cannabis percentages print(fuc2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md1 = nesarc.groupby('MAJORDEP12').size() # Major depression diagnoses counts print(md1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md2 = nesarc.groupby('MAJORDEP12').size() * 100 / len(nesarc) # Major depression diagnoses percentages print(md2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga1 = nesarc.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts print(ga1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga2 = nesarc.groupby('GENAXDX12').size() * 100 / len(nesarc) # Generalized anxiety diagnoses percentages print(ga2)
# Frequency distributions of major depression and general anxiety diagnoses variables for both last 12 months and prior cannabis users, ages 18-30 (subset1)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu1 = subsetc1.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset1) print(mdu1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu2 = subsetc1.groupby('MAJORDEP12').size() * 100 / len(subsetc1) # Major depression diagnoses percentages (subset1) print(mdu2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau1 = subsetc1.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset1) print(gau1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau2 = subsetc1.groupby('GENAXDX12').size() * 100 / len(subsetc1) # Generalized anxiety diagnoses percentages (subset1) print(gau2)
# Frequency distributions of major depression and general anxiety diagnoses variables of non-users, ages 18-30 (subset2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn1 = subsetc2.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset2) print(mdn1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn2 = subsetc2.groupby('MAJORDEP12').size() * 100 / len(subsetc2) # Major depression diagnoses percentages (subset2) print(mdn2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan1 = subsetc2.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset2) print(gan1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan2 = subsetc2.groupby('GENAXDX12').size() * 100 / len(subsetc2) # Generalized anxiety diagnoses percentages (subset2) print(gan2)
###################################################################################################################################
# Quartile age split, cut function, 4 groups (18-21, 21-24, 24-27, 27-30)
subsetc3['AGE4GROUPS'] = pandas.qcut(subsetc3.AGE, 4, labels=["1=17-21","2=21-24","3=24-27","4=27-30"])
print("Counts for age splitted in 4 groups: 18-21, 21-24, 24-27, 27-30") age4g1 = subsetc3.groupby('AGE4GROUPS').size() print(age4g1)
print("Percentages for age splitted in 4 groups: 18-21, 21-24, 24-27, 27-30") age4g2 = subsetc3.groupby('AGE4GROUPS').size() * 100 / len(subsetc3) print(age4g2)
print("Counts of observations within each of the age group four categories") subsetc3['AGE4GROUPS'] = pandas.cut(subsetc3.AGE, [17, 21, 24, 27, 30]) print (pandas.crosstab(subsetc3['AGE4GROUPS'], subsetc3['AGE']))
# Frequency distribution of cannabis use variable for ages 18-30 (subset3) with 9 set to NaN, number of missing data
subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].replace(99, numpy.nan)
recode = {1: 1, 2: 2, 9: "NaN"} subsetc3['CUMD'] = subsetc3['S3BQ1A5'].map(recode)
print("Counts for cannabis use ages 18-30 with missing data set to NaN, variable S3BQ1A5 - Section 3B") cuy1 = subsetc3.groupby('CUMD').size() # Cannabis use counts (subset3) print(cuy1) print("Percentages for cannabis use age 18-30 with missing data set to NaN, variable S3BQ1A5 - Section 3B") cuy2 = subsetc3.groupby('CUMD').size() * 100 / len(subsetc3) # Cannabis use percentages (subset3) print(cuy2)
# Frequency distribution of monthly average cannabis used (when using the most) variable for ages 18-30 (subset5)
recode1 = {1: 10, 2: 9, 3: 8, 4: 7, 5: 6, 6: 5, 7: 4, 8: 3, 9: 2, 10: 1} # Dictionary with details of frequency variable reverse-recode subsetc5['CUFREQ'] = subsetc5['S3BD5Q2E'].map(recode1) # Change variable name from S3BD5Q2E to CUFREQ recode2 = {1: 30, 2: 25, 3: 14, 4: 6, 5: 3, 6: 1, 7: 0.8, 8: 0.5, 9: 0.3, 10: 0.1} # Monthly average cannabis used subsetc5['CUFREQMO'] = subsetc5['S3BD5Q2E'].map(recode2) # Change variable name from S3BD5Q2E to CUFREQMO
print("Counts for average cannabis used per month when using the most, variable CUFREQMO") fucy1 = subsetc5.groupby('CUFREQMO').size() # Frequency of used cannabis counts (subset5) print(fucy1) print("Percentages for average cannabis used per month when using the most, variable CUFREQMO") fucy2 = subsetc5.groupby('CUFREQMO').size() * 100 / len(subsetc5) # Frequency of used cannabis percentages (subset5) print(fucy2)
# Secondary variable creation, NUMJOPMOTH_EST, number of joints per month
subsetc5['NUMJOPMOTH_EST'] = subsetc5['CUFREQMO'] * subsetc5['S3BQ4'] subsetc4 = subsetc5[['IDNUM' , 'S3BQ4' , 'CUFREQMO' , 'NUMJOPMOTH_EST']] head30 = subsetc4.head(30) print("Number of cannabis joints smoked per month when using the most, first 30 observations, new variable NUMJOPMOTH_EST") print(head30)
# Frequency distribution for both major depression and general anxiety diagnoses, new variable cration NUMMDGENANX
subsetc1['NUMMDGENANX'] = subsetc1['MAJORDEP12'] + subsetc1['GENAXDX12']
print("Counts for major depression and general anxiety diagnoses in cannabis users, ages 18-30, variable NUMMDGENANX") ndg1 = subsetc1.groupby('NUMMDGENANX').size() print(ndg1) print("Percentages for major depression and general anxiety diagnoses in cannabis users, ages 18-30, variable NUMMDGENANX") ndg2 = subsetc1.groupby('NUMMDGENANX').size() * 100 / len(subsetc1) print(ndg2)
# Newly managed depression and anxiety variables, in cannabis users ages 18-30, both last 12 months and prior (subset1), define function
def DEPRESSIONANXIETY (row): if row['NUMMDGENANX'] == 0 : return 0 if row['NUMMDGENANX'] > 1 : return 1 if row['MAJORDEP12'] == 1 : return 2 if row['GENAXDX12'] == 1 : return 3
subsetc1['DEPRESSIONANXIETY'] = subsetc1.apply (lambda row: DEPRESSIONANXIETY (row), axis=1) subsetc6 = subsetc1[['IDNUM' , 'MAJORDEP12' , 'GENAXDX12' , 'NUMMDGENANX' , 'DEPRESSIONANXIETY']].copy() first30 = subsetc6.head(30) print("Depression and anxiety diagnoses counts for cannabis users in last 12 months and prior, ages 18-30, new variable DEPRESSIONANXIETY") print(first30)
####################################################################################################################################
print("Counts for average number of joints smoked per month, ages 18-30, variable NUMJOPMOTH_EST") njpm1 = subsetc5.groupby('NUMJOPMOTH_EST').size() print(njpm1) print("Percentages for average number of joints smoked per month, ages 18-30, variable NUMJOPMOTH_EST") njpm2 = subsetc5.groupby('NUMJOPMOTH_EST').size() * 100 / len(subsetc5) print(njpm2)
# Change format of S3BQ1A5, S3BD5Q2E, MAJORDEP12, GENAXDX12 to categorical
subsetc3['S3BQ1A5'] = subsetc3['S3BQ1A5'].astype('category') subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].astype('category') subsetc1['MAJORDEP12'] = subsetc1['MAJORDEP12'].astype('category') subsetc1['GENAXDX12'] = subsetc1['GENAXDX12'].astype('category')
# Change the numbers with strings, rename x-axis categories
subsetc1['MAJORDEP12'] = subsetc1['MAJORDEP12'].cat.rename_categories(["No","Yes"]) subsetc1['GENAXDX12'] = subsetc1['GENAXDX12'].cat.rename_categories(["No","Yes"]) subsetc3['S3BQ1A5'] = subsetc3['S3BQ1A5'].cat.rename_categories(["Yes","No","Unknown"]) subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].cat.rename_categories(["Every day","Nearly every day","3-4 times/week","1-2 times/week","2-3 times/month","Once a month","7-11 times/year","3-6 times/year","2 times/year","Once a year"])
# Univariate bar chart for categorical variables, S3BQ1A5, S3BD5Q2E, MAJORDEP12, GENAXDX12, which stand for cannabis use, frequency of use, major depression and general anxiety
seaborn.countplot(x='S3BQ1A5', data=subsetc3) plt.xlabel('Cannabis use in ages 18-30') plt.title('Cannabis use, ages 18-30') plt.show() print ('Describe cannabis use variable') djpm2 = subsetc3['S3BQ1A5'].describe() print (djpm2)
plt.figure(figsize=(12,4)) # Change plot size ax5 = seaborn.countplot(x='S3BD5Q2E', data=subsetc5) ax5.set_xticklabels(ax5.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use in ages 18-30') plt.title('Frequency of cannabis use, ages 18-30') plt.show() print ('Describe frequency of cannabis use variable') djpm5 = subsetc5['S3BD5Q2E'].describe() print (djpm5)
seaborn.countplot(x='MAJORDEP12', data=subsetc1) plt.xlabel('Major depression diagnoses in last 12 months') plt.title('Major depression diagnosed in cannabis users (both last 12 months and prior), ages 18-30') plt.show() print ('Describe major depression variable') djpm3 = subsetc1['MAJORDEP12'].describe() print (djpm3) seaborn.countplot(x='GENAXDX12', data=subsetc1) plt.xlabel('General anxiety diagnoses in last 12 months') plt.title('General anxiety diagnosed in cannabis users (both last 12 months and prior), ages 18-30') plt.show() print ('Describe general anxiety variable') djpm4 = subsetc1['GENAXDX12'].describe() print (djpm4)
# Univariate bar chart for quantitative variable, NUMJOPMOTH_EST, that stands for the number of joints smoked per month when using the most, ages 18-30
plt.figure(figsize=(10,4)) # Change plot size seaborn.distplot(subsetc5["NUMJOPMOTH_EST"].dropna(), kde=False) plt.xlabel('Number of joints smoked per month') plt.title('Estimated number of joints smoked per month by cannabis users, ages 18-30') plt.show()
# Center and spread measurements
print ('Spread') std1 = subsetc5['NUMJOPMOTH_EST'].std() print (std1)
print ('Mode') mode1 = subsetc5['NUMJOPMOTH_EST'].mode() print (mode1)
print ('Mean') mean1 = subsetc5['NUMJOPMOTH_EST'].mean() print (mean1)
print ('Median') median1 = subsetc5['NUMJOPMOTH_EST'].median() print (median1)
# Plot for groups of total number of joints smoked per month, variable NUMJOPMOTH_EST
subsetc5['NUMJOPMOTH_EST'] = pandas.cut(subsetc5.NUMJOPMOTH_EST, [0, 1, 10, 20, 30, 50, 70, 90, 110, 130, 150, 200, 250, 300, 2970]) # Split the number into groups subsetc5['NUMJOPMOTH_EST'] = subsetc5['NUMJOPMOTH_EST'].astype('category') # Rename x-axis categories of the plot subsetc5['NUMJOPMOTH_EST'] = subsetc5['NUMJOPMOTH_EST'].cat.rename_categories(["<1","1-10","11-20","21-30","31-50","51-70","71-90","90-110","111-130","131-150","151-200","201-250","251-300",">300"]) plt.figure(figsize=(10,4)) # Change plot size ax = seaborn.countplot(x='NUMJOPMOTH_EST', data=subsetc5) ax.set_xticklabels(ax.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Number of joints smoked per month') plt.title('Estimated number of joints smoked per month by cannabis users, ages 18-30') plt.show()
print ('Describe number of joints smoked per month') djpm1 = subsetc5['NUMJOPMOTH_EST'].describe() print (djpm1)
# Bivariate bar graph C->Q, major depression as response variable and number of joints smoked per month as explanatory variable
nesarc['MAJORDEP12'] = nesarc['MAJORDEP12'].convert_objects(convert_numeric=True)
plt.figure(figsize=(10,4)) # Change plot size ax1 = seaborn.factorplot(x="NUMJOPMOTH_EST", y="MAJORDEP12", data=subsetc5, kind="bar", ci=None) ax1.set_xticklabels(ax.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Joints smoked per Month') plt.ylabel('Proportion of major depression') plt.show()
# Bivariate bar graph C->Q, general anxiety as response variable and number of joints smoked per month as explanatory variable
nesarc['GENAXDX12'] = nesarc['GENAXDX12'].convert_objects(convert_numeric=True)
plt.figure(figsize=(10,4)) # Change plot size ax2 = seaborn.factorplot(x="NUMJOPMOTH_EST", y="GENAXDX12", data=subsetc5, kind="bar", ci=None) ax2.set_xticklabels(ax.get_xticklabels(), rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Joints smoked per Month') plt.ylabel('Proportion of general anxiety') plt.show()
nesarc['S3BQ1A5'] = nesarc['S3BQ1A5'].convert_objects(convert_numeric=True) subsetc3['S3BQ1A5']=subsetc3['S3BQ1A5'].replace(9, numpy.nan)
# Bivariate bar graph C->C, major depression as response variable and cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(10,4)) # Change plot size seaborn.factorplot(x="S3BQ1A5", y="MAJORDEP12", data=subsetc3, kind="bar", ci=None) plt.xlabel('Cannabis use ages 18-30') plt.ylabel('Proportion of major depression') plt.show()
# Bivariate bar graph C->C, general anxiety as response variable and cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(10,4)) # Change plot size seaborn.factorplot(x="S3BQ1A5", y="GENAXDX12", data=subsetc3, kind="bar", ci=None) plt.xlabel('Cannabis use ages 18-30') plt.ylabel('Proportion of general anxiety') plt.show()
# Change frequency variable type to categorical
subsetc5['S3BD5Q2E'] = subsetc5['S3BD5Q2E'].astype('category')
# Bivariate bar graph C->C, major depression as response variable and frequency of cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(12,4)) # Change plot size ax3 = seaborn.factorplot(x="S3BD5Q2E", y="MAJORDEP12", data=subsetc5, kind="bar", ci=None) ax3.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use ages 18-30') plt.ylabel('Proportion of major depression') plt.show()
# Bivariate bar graph C->C, general anxiety as response variable and frequency of cannabis use in ages 18 to 30 as explanatory variable
plt.figure(figsize=(12,4)) # Change plot size ax4 = seaborn.factorplot(x="S3BD5Q2E", y="GENAXDX12", data=subsetc5, kind="bar", ci=None) ax4.set_xticklabels(rotation=40, ha="right") # X-axis labels rotation plt.xlabel('Frequency of cannabis use ages 18-30') plt.ylabel('Proportion of general anxiety') plt.show()
## Output Discussion with links:
# Making Data Management Decisions
## Preview In this assignment you will find three data managed variables, provided from NESARC codebook, as frequency distributions. For the variable **‘AGE’**, I decided to split the ages of the observations in four categories (18-21, 21-24, 24-27, 27-30), in order to examine the age frequency distribution of the sample into quartiles. Furthermore, I included missing data in the cannabis use variable distribution for ages 18 to 30, with the row ‘Unknown’ set to ‘nan’. In addition, after recoding variable **‘S3BD5Q2E’**, which represents average rate of cannabis use, I created a new variable called **‘CUFREQMO’** that indicates how many days an individual used cannabis per month, when using the most. Moreover, once I ran frequency distribution for the variable **‘CUFREQMO’** and calculated the counts and the percentages of this use per month, I created a secondary variable with the name **‘NUMJOPMOTH_EST’**. This variable estimates the quantity of cannabis joints smoked per month for the first 30 participants (18-30) when using the most, by multiplying the amount of joints smoked per day (variable **‘S3BQ4’**) with the variable **‘CUFREQMO’**, which indicates the number of days an individual smoked per month. I finished with the creation of one more variable called **‘NUMMDGENANX’** that stands for the total number of participants who were diagnosed with either major depression, general anxiety or both illnesses. After examining the frequency distribution of this variable, I used the define function to create one last variable with the name **‘DEPRESSIONANXIETY’**, which characterizes the mental state of the first 30 random cannabis users aged 18 to 30, as far as these two disorders are concerned. For the code and the output i used Spyder (IDE).
## Output 
Firstly, a new variable called **‘AGE4GROUPS’** was necessary to be created in order to divide participants, aged between 18 and 30 years old, into four age groups: 18-21, 21-24, 24-27 and 27-30. From the frequency distribution it can be noticed that about 30.73% were between 18 and 21 years old, 23.78% were between 21 and 24, 20.65% fell into 24-27 group and 24.82% fell into 27-30 group.

For the frequency distribution of variable **‘S3BQ1A5′** the missing data is included and option 9 is set to ‘NaN’, which represents the choice ‘Unknown’. As we can see the number of the missing data is 81 individuals and the proportion is 0.84% which is significantly small. Regarding the new variable with the name **’CUFREQMO’** which I created, the distribution results show that about 15.96% smoked cannabis less than once per year, whereas approximately 22.13% used to smoke cannabis in an everyday basis. The rest of the cannabis use rates vary from about 2.65% for less than once per month, to 11.6% for an average use of 6 times per month.

Taking into account the new variable **‘NUMJOPMOTH_EST’** which I created, the output illustrates the estimation of the quantity of joints smoked by young adults aged 18 to 30 per month, by multiplying the quantity of joints smoked per day with the total number of days that they used cannabis per month. By examining the first 30 observations of the sample, it is noticeable that the majority of individuals smoked approximately less than 10 joints per month. On the other hand there were also some participants who were smoking more than 100 joints monthly, since individuals with ID numbers 511 and 506 for example, peaked at 180 and 150 joints per month respectively.

As far as the new secondary variable **‘NUMMDGENANX’** is concerned, it is obvious that around 77% of cannabis users aged between 18 and 30 were mentally healthy in the last 12 months, after using marijuana for more than 1 year, while a percentage of 21.07% were diagnosed with either major depression or general anxiety. Only 1.91% of them were diagnosed with both mental disorders mentioned above. Finally, regarding the first 30 observations table, presented in the end, the define function used, returns 0 if an individual had no problem with either depression or anxiety in the last 12 months, 1 if an individual met the criteria for both disorders, 2 if an individual was diagnosed only with major depression and 3 if an individual was diagnosed only with general anxiety. For the results, a new variable named **‘DEPRESSIONANXIETY’** was created.
0 notes
Text
Assignment, Week 2
## Code:
import pandas import numpy nesarc = pandas.read_csv ("nesarc_pds.csv" , low_memory=False) # load NESARC dataset nesarc.columns = map(str.upper , nesarc.columns) pandas.set_option('display.float_format' , lambda x:'%f'%x) print (len(nesarc)) # Number of observations print (len(nesarc.columns)) # Number of variables
# Change my variables to numeric
nesarc['AGE'] = nesarc['AGE'].convert_objects(convert_numeric=True) nesarc['S3BQ1A5'] = nesarc['S3BQ1A5'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2B'] = nesarc['S3BD5Q2B'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2E'] = nesarc['S3BD5Q2E'].convert_objects(convert_numeric=True) nesarc['MAJORDEP12'] = nesarc['MAJORDEP12'].convert_objects(convert_numeric=True) nesarc['GENAXDX12'] = nesarc['GENAXDX12'].convert_objects(convert_numeric=True)
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BD5Q2B']==3) & (nesarc['S3BQ1A5']==1)] # Cannabis users both last 12 months and prior, ages 18-30 subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==2)] # Non-users, ages 18-30 subsetc2 = subset2.copy()
subset3 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30 subsetc3 = subset3.copy()
# Frequency distributions of variables (groupby function) of the entire sample
print("Counts for cannabis use, variable S3BQ1A5 - Section 3B") cu1 = nesarc.groupby('S3BQ1A5').size() # Cannabis use counts print(cu1) print("Percentages for cannabis use, variable S3BQ1A5 - Section 3B") cu2 = nesarc.groupby('S3BQ1A5').size() * 100 / len(nesarc) # Cannabis use percentages print(cu2)
print("Counts for used cannabis in the last 12 months/prior to last 12 months/both time periods, variable S3BD5Q2B - Section 3B") uc1 = nesarc.groupby('S3BD5Q2B').size() # Used cannabis time periods counts print(uc1) print("Percentages for used cannabis in the last 12 months/prior to last 12 months/both time periods, S3BD5Q2B - Section 3B") uc2 = nesarc.groupby('S3BD5Q2B').size() * 100 / len(nesarc) # Used cannabis time periods percentages print(uc2)
print("Counts for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc1 = nesarc.groupby('S3BD5Q2E').size() # Frequency of used cannabis counts print(fuc1) print("Percentages for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc2 = nesarc.groupby('S3BD5Q2E').size() * 100 / len(nesarc) # Frequency of used cannabis percentages print(fuc2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md1 = nesarc.groupby('MAJORDEP12').size() # Major depression diagnoses counts print(md1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md2 = nesarc.groupby('MAJORDEP12').size() * 100 / len(nesarc) # Major depression diagnoses percentages print(md2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga1 = nesarc.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts print(ga1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga2 = nesarc.groupby('GENAXDX12').size() * 100 / len(nesarc) # Generalized anxiety diagnoses percentages print(ga2)
# Frequency distribution of cannabis use variable for ages 18-30 (subset3) print("Counts for cannabis use ages 18-30, variable S3BQ1A5 - Section 3B") cuy1 = subsetc3.groupby('S3BQ1A5').size() # Cannabis use counts (subset3) print(cuy1) print("Percentages for cannabis use age 18-30, variable S3BQ1A5 - Section 3B") cuy2 = subsetc3.groupby('S3BQ1A5').size() * 100 / len(subsetc3) # Cannabis use percentages (subset3) print(cuy2)
# Frequency distributions of major depression and general anxiety diagnoses variables for both last 12 months and prior cannabis users, ages 18-30 (subset1)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu1 = subsetc1.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset1) print(mdu1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu2 = subsetc1.groupby('MAJORDEP12').size() * 100 / len(subsetc1) # Major depression diagnoses percentages (subset1) print(mdu2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau1 = subsetc1.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset1) print(gau1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau2 = subsetc1.groupby('GENAXDX12').size() * 100 / len(subsetc1) # Generalized anxiety diagnoses percentages (subset1) print(gau2)
# Frequency distributions of major depression and general anxiety diagnoses variables of non-users, ages 18-30 (subset2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn1 = subsetc2.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset2) print(mdn1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn2 = subsetc2.groupby('MAJORDEP12').size() * 100 / len(subsetc2) # Major depression diagnoses percentages (subset2) print(mdn2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan1 = subsetc2.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset2) print(gan1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan2 = subsetc2.groupby('GENAXDX12').size() * 100 / len(subsetc2) # Generalized anxiety diagnoses percentages (subset2) print(gan2)
## Output:

## Describing the Output:
## Preview In order to examine frequency distributions for my selected variables, I chose to use Spyder, a python integrated development environment (IDE). Once I have run frequency distributions for all of my chosen variables, I decided to subset my dataset and ask my question based on a specific set of observations, instead of the entire sample. I found myself most interested in the association between cannabis use and major depression as well as general anxiety disorders diagnosed in the last 12 months, but only among young adults between 18 and 30 years old who have used cannabis both in last 12 months and before. Moreover, the results of the subset mentioned above will be compared with the outcomes of my secondary subset, which aims to examine the same association, but this time only among young adults, aged 18-30, who have never used cannabis. Finally, a general proportion of cannabis use among adults between 18 and 30 years old, will also be presented.
### Subset1 For the variable **‘AGE’** I decided to include two logic statements which are particular rows in the NESARC dataset. These are [‘AGE’]>=18 and ['AGE’]<=30. In addition, as far as variable **'S3BQ1A5’** is concerned, which represents cannabis use, the logic statement ['S3BQ1A5’]==1 (1=Yes) should also be included. Finally, for the variable **'S3BD5Q2B’** that represents the period of the use, only the row ['S3BD5Q2B’]==3 (3=During both time periods) was taken into account.
### Subset2 Same as subset1, the logic statements for variable **‘AGE’** are ['AGE’]>=18 and ['AGE’]<=30. Alternatively in this case, since we need to examine the results among only non-users young adults, the logic statement for cannabis use variable **'S3BQ1A5’** is ['S3BQ1A5’]==2 (2=No).
## Frequency Table 
or shown above
## Report A random sample of 9535 U.S. young adults, aged 18-30, were asked, as a part of NESARC survey, the following question: “Have you ever used cannabis?” A percentage of 25.29% answered “Yes”, whereas 73.85% answered “No”. Also a significantly small percentage of 0.84%, fell into category 9 (“Unknown“) which is our missing data.
Of the total number of participants (18-30) who answered “Yes” to the question of cannabis use, only those who were smoking marijuana in last 12 months and prior were taken into consideration for the next questions.
To the question of “Have you been diagnosed with non-hierarchical major depression in the last 12 months?”, about 20.95% of the cannabis users responded “Yes” while only 8.42% of the non-users answered the same.
For the question, ”Have you been diagnosed with non-hierarchical generalized anxiety in the last 12 months?”, a proportion of 3.95% of cannabis users were diagnosed positive to anxiety disorders compared to 1.63% of the non-users.
## Conclusion To sum up, looking through the the frequency table it can be noticed that there are some slight differences between the percentages of cannabis users compared to non-users. Major depression cases in cannabis users young adults (20.95%) seem to be slightly more than double compared to those of non-users (8.42%).In addition, general anxiety diagnoses in cannabis users (3.95%) appear to be also marginally more than double in comparison to the non-users (1.63%). It could be supported that there is a relative association between cannabis and such mental disorders, thus cannabis use increases the likelihood of meeting criteria for depression or general anxiety in the future. However, the sample is extremely small and it is unclear how representative it is, making the findings less reliable, since a large amount of error may be involved.
0 notes
Text
Week 2 Submission
My First Program - Python
Preview-
In order to examine frequency distributions for my selected variables, I chose to use Jupyter Lab. Once I have run frequency distributions for all of my chosen variables, I decided to subset my dataset and ask my question based on a specific set of observations, instead of the entire sample. I found myself most interested in the association between cannabis use and major depression as well as general anxiety disorders diagnosed in the last 12 months, but only among young adults between 18 and 30 years old who have used cannabis both in last 12 months and before. Moreover, the results of the subset mentioned above will be compared with the outcomes of my secondary subset, which aims to examine the same association, but this time only among young adults, aged 18-30, who have never used cannabis. Finally, a general proportion of cannabis use among adults between 18 and 30 years old, will also be presented.
My Program-
import pandas import numpy nesarc = pandas.read_csv ("nesarc_pds.csv" , low_memory=False) # load NESARC dataset nesarc.columns = map(str.upper , nesarc.columns) pandas.set_option('display.float_format' , lambda x:'%f'%x) print (len(nesarc)) # Number of observations print (len(nesarc.columns)) # Number of variables
# Change my variables to numeric
nesarc['AGE'] = nesarc['AGE'].convert_objects(convert_numeric=True) nesarc['S3BQ1A5'] = nesarc['S3BQ1A5'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2B'] = nesarc['S3BD5Q2B'].convert_objects(convert_numeric=True) nesarc['S3BD5Q2E'] = nesarc['S3BD5Q2E'].convert_objects(convert_numeric=True) nesarc['MAJORDEP12'] = nesarc['MAJORDEP12'].convert_objects(convert_numeric=True) nesarc['GENAXDX12'] = nesarc['GENAXDX12'].convert_objects(convert_numeric=True)
subset1 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BD5Q2B']==3) & (nesarc['S3BQ1A5']==1)] # Cannabis users both last 12 months and prior, ages 18-30 subsetc1 = subset1.copy()
subset2 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30) & (nesarc['S3BQ1A5']==2)] # Non-users, ages 18-30 subsetc2 = subset2.copy()
subset3 = nesarc[(nesarc['AGE']>=18) & (nesarc['AGE']<=30)] # Ages 18-30 subsetc3 = subset3.copy()
# Frequency distributions of variables (groupby function) of the entire sample
print("Counts for cannabis use, variable S3BQ1A5 - Section 3B") cu1 = nesarc.groupby('S3BQ1A5').size() # Cannabis use counts print(cu1) print("Percentages for cannabis use, variable S3BQ1A5 - Section 3B") cu2 = nesarc.groupby('S3BQ1A5').size() * 100 / len(nesarc) # Cannabis use percentages print(cu2)
print("Counts for used cannabis in the last 12 months/prior to last 12 months/both time periods, variable S3BD5Q2B - Section 3B") uc1 = nesarc.groupby('S3BD5Q2B').size() # Used cannabis time periods counts print(uc1) print("Percentages for used cannabis in the last 12 months/prior to last 12 months/both time periods, S3BD5Q2B - Section 3B") uc2 = nesarc.groupby('S3BD5Q2B').size() * 100 / len(nesarc) # Used cannabis time periods percentages print(uc2)
print("Counts for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc1 = nesarc.groupby('S3BD5Q2E').size() # Frequency of used cannabis counts print(fuc1) print("Percentages for frequency of used cannabis when using the most, variable S3BD5Q2E - Section 3B") fuc2 = nesarc.groupby('S3BD5Q2E').size() * 100 / len(nesarc) # Frequency of used cannabis percentages print(fuc2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md1 = nesarc.groupby('MAJORDEP12').size() # Major depression diagnoses counts print(md1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months, variable MAJORDEP12 - Section 14") md2 = nesarc.groupby('MAJORDEP12').size() * 100 / len(nesarc) # Major depression diagnoses percentages print(md2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga1 = nesarc.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts print(ga1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months, variable GENAXDX12 - Section 14") ga2 = nesarc.groupby('GENAXDX12').size() * 100 / len(nesarc) # Generalized anxiety diagnoses percentages print(ga2)
# Frequency distribution of cannabis use variable for ages 18-30 (subset3) print("Counts for cannabis use ages 18-30, variable S3BQ1A5 - Section 3B") cuy1 = subsetc3.groupby('S3BQ1A5').size() # Cannabis use counts (subset3) print(cuy1) print("Percentages for cannabis use age 18-30, variable S3BQ1A5 - Section 3B") cuy2 = subsetc3.groupby('S3BQ1A5').size() * 100 / len(subsetc3) # Cannabis use percentages (subset3) print(cuy2)
# Frequency distributions of major depression and general anxiety diagnoses variables for both last 12 months and prior cannabis users, ages 18-30 (subset1)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu1 = subsetc1.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset1) print(mdu1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable MAJORDEP12 - Section 14") mdu2 = subsetc1.groupby('MAJORDEP12').size() * 100 / len(subsetc1) # Major depression diagnoses percentages (subset1) print(mdu2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau1 = subsetc1.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset1) print(gau1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (both last 12 months and prior cannabis users, ages 18-30), variable GENAXDX12 - Section 14") gau2 = subsetc1.groupby('GENAXDX12').size() * 100 / len(subsetc1) # Generalized anxiety diagnoses percentages (subset1) print(gau2)
# Frequency distributions of major depression and general anxiety diagnoses variables of non-users, ages 18-30 (subset2)
print("Counts for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn1 = subsetc2.groupby('MAJORDEP12').size() # Major depression diagnoses counts (subset2) print(mdn1) print("Percentages for non-hierarchical major depression diagnoses in last 12 months (non-users, ages 18-30), variable MAJORDEP12 - Section 14") mdn2 = subsetc2.groupby('MAJORDEP12').size() * 100 / len(subsetc2) # Major depression diagnoses percentages (subset2) print(mdn2)
print("Counts for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan1 = subsetc2.groupby('GENAXDX12').size() # Generalized anxiety diagnoses counts (subset2) print(gan1) print("Percentages for non-hierarchical generalized anxiety diagnoses in last 12 months (non-users, ages 18-30), variable GENAXDX12 - Section 14") gan2 = subsetc2.groupby('GENAXDX12').size() * 100 / len(subsetc2) # Generalized anxiety diagnoses percentages (subset2) print(gan2)
OUTPUT OF THE FOLLOWING CODE-

Subset1 For the variable **‘AGE’** I decided to include two logic statements which are particular rows in the NESARC dataset. These are [‘AGE’]>=18 and ['AGE’]<=30. In addition, as far as variable **'S3BQ1A5’** is concerned, which represents cannabis use, the logic statement ['S3BQ1A5’]==1 (1=Yes) should also be included. Finally, for the variable **'S3BD5Q2B’** that represents the period of the use, only the row ['S3BD5Q2B’]==3 (3=During both time periods) was taken into account.
Subset2 Same as subset1, the logic statements for variable **‘AGE’** are ['AGE’]>=18 and ['AGE’]<=30. Alternatively in this case, since we need to examine the results among only non-users young adults, the logic statement for cannabis use variable **'S3BQ1A5’** is ['S3BQ1A5’]==2 (2=No).
Report A random sample of 9535 U.S. young adults, aged 18-30, were asked, as a part of NESARC survey, the following question: “Have you ever used cannabis?” A percentage of 25.29% answered “Yes”, whereas 73.85% answered “No”. Also a significantly small percentage of 0.84%, fell into category 9 (“Unknown“) which is our missing data.
Of the total number of participants (18-30) who answered “Yes” to the question of cannabis use, only those who were smoking marijuana in last 12 months and prior were taken into consideration for the next questions.
To the question of “Have you been diagnosed with non-hierarchical major depression in the last 12 months?”, about 20.95% of the cannabis users responded “Yes” while only 8.42% of the non-users answered the same.
For the question, ”Have you been diagnosed with non-hierarchical generalized anxiety in the last 12 months?”, a proportion of 3.95% of cannabis users were diagnosed positive to anxiety disorders compared to 1.63% of the non-users.
Conclusion To sum up, looking through the the frequency table it can be noticed that there are some slight differences between the percentages of cannabis users compared to non-users. Major depression cases in cannabis users young adults (20.95%) seem to be slightly more than double compared to those of non-users (8.42%).In addition, general anxiety diagnoses in cannabis users (3.95%) appear to be also marginally more than double in comparison to the non-users (1.63%). It could be supported that there is a relative association between cannabis and such mental disorders, thus cannabis use increases the likelihood of meeting criteria for depression or general anxiety in the future. However, the sample is extremely small and it is unclear how representative it is, making the findings less reliable, since a large amount of error may be involved.
0 notes