#embryonic genome editing
Explore tagged Tumblr posts
Text
Latest Advances in Gene and Cell Therapies Transform Healthcare

Gene and cell therapies represent a ground-breaking advancement in medical science, offering potential cures for a variety of previously untreatable diseases. These therapies are revolutionizing how we provide targeted healthcare by modifying genetic material or using cells to restore or alter biological functions. Early interventions in congenital disorders can significantly reduce long-term health complications, offering a healthier start to life for newborns. Thus, the potential of gene and cell therapies to transform medical treatments is immense, especially in the field of natal and prenatal care.
A notable example of gene therapy involved the birth of the first babies with edited genes. In 2018, Dr. Jiankui announced the birth of twin girls whose genes were edited using CRISPR technology. He edited and deactivated a gene known as CCR5 with the goal of conferring resistance to HIV in those girls.
Latest Developments in Gene and Cell Therapies
The field of gene and cell therapies is crucial in the mainstream as drug-regulating authorities approve treatments for diseases like lymphoma and muscular dystrophy. Let us explore the latest developments regarding these therapies.
Non-Hodgkin lymphoma (NHL) accounts for about 4% of all cancers in the US, with an estimated 80,620 new cases expected this year. In this regard, Bristol Myers Squibb’s Breyanzi, a CAR T cell therapy, was approved in 2024 by the FDA, which utilizes the patient’s immune system to target and destroy cancer cells.
In 2024, the FDA approved Sarepta Therapeutics’ Elevidys, a gene therapy for Duchenne muscular dystrophy (DMD), which affects approximately 1 in 3,500 to 5000 male births worldwide, typically manifesting between ages 3 and 6. This groundbreaking offers new hope by addressing the root cause of this debilitating condition.
Exploring Current and Future Applications
CRISPR and Genome Editing: CRISPR technology has revolutionized genome editing, offering precise modifications to DNA and correcting genetic defects at their source. This technology is being explored for a variety of applications including current and future applications. However, acquiring approvals to run trials on humans has always been challenging, yet the CTX001 stands out with its success in this regard. The CTX001 is an autologous gene-edited stem cell therapy developed by CRISPR Therapeutics and Vertex Pharmaceuticals.
Dr. Haydar Frangoul, the medical director at HCA Sarah Cannon Research Institute Center, has been treating the first patient in the CTX001 trial for SCD therapy. The patient had battled sickle cell disease for 34 years before undergoing this one-time treatment. Post-treatment, her blood showed a significant proportion of fetal hemoglobin levels, enabling her to avoid blood transfusions and pain attacks without major side effects.
Stem Cell Research: These cells have the unique ability to differentiate into various cell types, making them invaluable for regenerative medicine. Research in stem cell therapy aims to treat conditions such as Parkinson’s disease, diabetes, and spinal cord injuries by replacing damaged cells with healthy ones in the near future. A notable example is a study using device-encapsulated pancreatic precursor cells derived from human embryonic stem cells. This study has shown that increased cell doses in optimized devices lead to detectable insulin production and improved glucose control.
CAR-T Cell Therapy: This therapy has shown impressive results in treating certain types of leukemia and lymphoma, offering hope for patients who have not responded to traditional treatments. This innovative approach uses modified T-cells to target and kill cancer cells. The future of CAR-T therapy looks promising, thereby expanding its application to treat more types of cancers, including solid tumors.
Gene Silencing and RNA-based Therapies: Emerging technologies like RNA interference (RNAi) and antisense oligonucleotides (ASOs) are being developed to silence harmful genes. An RNAi therapy like ‘AMVUTTRA’ developed by Alnylam, is approved in the US for treating polyneuropathy of hereditary transthyretin-mediated (hATTR) amyloidosis in adults. Thus, the future use of RNA therapies includes the treatment of neurodegenerative diseases like Huntington’s disease.
Understanding Ethical Considerations & the Role of Regulatory Bodies
Ethical frameworks must evolve amidst the concerns regarding ‘designer babies’, where genetic modifications used to select desired traits pose significant ethical dilemmas. A prominent example is the controversy of using CRISPR technology in human embryos, who claimed to have created the first gene-edited babies, sparking ethical debates and leading to his imprisonment. Several studies emphasize the importance of international regulatory standards and effective governance to ensure the responsible use of gene editing technologies.
Amidst the rapid pace of technological advancement, regulating gene and cell therapies needs rigorous safety standards. The regulatory bodies and agencies like the FDA’s Center for Biologics Evaluation and Research (CBER) in the US and the European Medicines Agency (EMA) in the EU play a critical role. Their frameworks include guidelines for approval of regenerative medicines and conditional or time-limited authorizations to facilitate quicker access to innovative treatments.
What the future beholds?
The future of gene and cell therapies lies in their integration into personalized medicine based on the genetic makeup of individual patients. Companies like CRISPR Therapeutics, Editas Medicine, and Intellia Therapeutics are at the forefront of research, developing therapies that could revolutionize the treatment of genetic disorders. As these therapies become more refined and accessible, they could significantly extend healthy life spans and improve the quality of life for millions.
#Gene and Cell Therapies#healthcare#lifesciences#genome editing#CRISPR technology#Stem Cell therapy#triton market research#market research reports
0 notes
Link
0 notes
Text
genome editing talen
As effective molecular scissors, TALENs are widely used to induce a variety of specific and efficient genomic modifications in various cell types and different model organisms. With technology development, TALs can be engineered to deliver virtually any enzyme to any site and perform gene-editing functions, including gene knockout, knockin, mutagenesis, gene tagging, and for disease and cell therapeutics models. It is documented that TALENs have been used to engineer stably modified human embryonic stem cell and induced pluripotent stem cell (IPSCs), moreover, they have great promises in the cell therapy of cancers and genetic disorders, such as sickle cell disease and xeroderma pigmentosum. Learn more about genome editing talen.
0 notes
Text
The ethical implications of new clinical embryology technologies

Embryology, a fascinating field of science that explores the early stages of human life, has witnessed a remarkable transformation due to advancements in technology. In this article, we delve into the ethical dimensions surrounding these new clinical embryology technologies, considering the importance of ethical practices and their integration into education and training. We also shed light on the role of SEART, a leading name in this domain, in ensuring ethical clinical embryology practices.
1. Introduction to Clinical Embryology
Clinical embryology is a pivotal field encompassing the study of early human development, playing a critical role in assisted reproductive technologies and research. This science explores the stages from fertilization to the development of a viable fetus. With the advent of new technologies, such as in vitro fertilization (IVF), pre-implantation genetic testing (PGT), and gene editing, clinical embryology has evolved significantly.
Definition and Significance
Clinical embryology involves the laboratory-based techniques and procedures utilized in assisted reproductive technologies and embryonic research. It holds immense significance in addressing fertility issues and advancing genetic research.
Evolution of Clinical Embryology
Over the years, clinical embryology has progressed from basic observation to sophisticated genetic manipulations, enabling better understanding and intervention in human reproduction.
Overview of New Technologies
Technological advancements, such as PGT and gene editing tools like CRISPR-Cas9, have revolutionized clinical embryology, offering unparalleled opportunities and raising ethical concerns.
2. The Rise of Clinical Embryology Programs
As the field expands, so does the need for skilled embryologists. Various educational programs cater to this demand, providing aspiring embryologists with the necessary knowledge and expertise.
Education and Training
Embryologist training involves a deep dive into the intricacies of human embryonic development, laboratory techniques, and ethical practices. Institutions globally offer specialized courses like Masters in Clinical Embryology and PG Diploma in Clinical Embryology.
Popular Courses and Specializations
Institutions provide specialized programs like "MSc in Clinical Embryology" and "Masters in Clinical Embryology," equipping students with the expertise required in the field.
Key Institutions Offering Programs
Renowned institutions like SEART offer comprehensive courses, integrating theoretical knowledge with hands-on experience, ensuring the ethical practice of clinical embryology.
3. Exploring Ethical Dimensions
The rapid advancement of clinical embryology technologies necessitates a thorough examination of the ethical implications associated with these developments.
Balancing Progress and Ethical Considerations
Striking a balance between scientific progress and ethical considerations is paramount to ensure the responsible and beneficial application of emerging technologies.
Ethical Concerns with Clinical Embryology Technologies
Issues like genetic screening, gene editing, and the creation of designer babies raise valid ethical concerns related to the alteration of the human genome and the potential misuse of technology.
Regulatory Frameworks and Guidelines
Adhering to established regulations and guidelines is vital in ensuring that clinical embryology technologies are utilized ethically and responsibly.
Stay tuned for the continuation of this article, where we'll discuss the crucial role that SEART plays in promoting ethical practices within the realm of clinical embryology.
We'll explore SEART's commitment to ethical standards and how they integrate ethics into their education and training programs. Plus, we'll share insightful case studies and testimonials emphasizing the ethical core of SEART's approach. Embark with us on this enlightening journey through the ethical landscape of clinical embryology!
#ClinicalEmbryology#EmbryologistTraining#PGDiplomaInEmbryology#MasterInEmbryology#EthicalEmbryology#SEART#MScClinicalEmbryology#ClinicalEmbryologyCourses#GeneticScreening
0 notes
Text
Introducing IASRM's Premiere Fellowship
Dive into the world of Stem Cell-Based Regenerative & Cellular Medicine with our comprehensive 1-year fellowship program. Recognizing the evolving needs of the medical community, we've crafted a curriculum that offers an in-depth understanding of stem cell therapy, regenerative medicine, and the most recent advancements in clinical research. 🔍 What Does Our Program Offer?
E-Learning Sessions: Convenient online lectures available through the IASRM LMS, ensuring flexibility for professionals. Hands-On Experiences: Learn practically with live demonstrations and hands-on training in New Delhi or our affiliated IASRM centers.
We provide multiple schedules throughout the year for your convenience. Thesis Submission: An opportunity to delve deep into a topic of choice, providing a platform for in-depth exploration and contribution to the field.
📘 Key Curriculum Highlights:
Foundations of Stem Cells & Their Clinical Applications
Exploring Embryonic Stem Cells, IPSC’s, and Ongoing Clinical Research
Basics & Applications of Platelet Rich Plasma (PRP)
Insights into BMAC-Bone, Haematopoietic Stem Cell Transplants, & Mesenchymal Stem Cells
A deep dive into Exosomes, Liposomes, and other regenerative therapies.
Ethical & Regulatory Requirements, Clinical Trials, and Translational Medicine insights
Commercial Aspects: Intellectual Property, Funding, & Marketing in Regenerative Medicine
Advanced topics like CAR-T Therapy, Genome Editing Tools, Tissue Engineering, and more.
Get ready to embark on a transformative journey, enhancing your knowledge, skills, and understanding of this pivotal field in medicine.
#iasrm #prabhumishra #IASRMDeepDive #StemCellMastery #RegenerativeMedicineCourse #HandsOnIASRM #PRPandBeyond #MedicalInnovation #CARTherapyInsights #FutureMedicineTraining #BiotechExplained #MedicineReimagined #IASRM2023
#prabhumishra#iasrm#IASRMDeepDive#StemCellMastery#RegenerativeMedicineCourse#HandsOnIASRM#PRPandBeyond#MedicalInnovation#CARTherapyInsights#FutureMedicineTraining#BiotechExplained#MedicineReimagined#IASRM2023
0 notes
Text
Fwd: Postdoc: CornellU.EvolutionOfNovelty
Begin forwarded message: > From: [email protected] > Subject: Postdoc: CornellU.EvolutionOfNovelty > Date: 11 March 2023 at 06:54:10 GMT > To: [email protected] > > > > Postdoctoral Research Associate Studying Evolution of Novelty > > Department of Ecology & Evolutionary Biology > > Cornell University, Ithaca, NY > > The College of Arts & Sciences at Cornell University embraces diversity > and seeks candidates who will contribute to a climate that supports > students, faculty, and staff of all identities and backgrounds. We > strongly encourage individuals from underrepresented and/or marginalized > identities to apply. > > Position Function > > The Babonis Lab in the Department of Ecology and Evolutionary Biology > at Cornell University seeks to hire a highly motivated postdoc to work > on the evolution of cnidocyte (stinging cell) diversity in cnidarians > (corals, sea anemones, jellyfish). The Babonis Lab is broadly interested > in understanding the origin of taxon-specific traits (novelties) across > levels of organization. > > Primary areas of interest in the lab include: the functional > diversification of cnidocytes (stinging cells), the emergence of > specialized animal secretory cell types, the evolution of regeneration > and stem cell identity, the origin of novel genes, and the assembly of > novel gene regulatory networks. > > The successful candidate will be expected to use a broad range of > cellular, molecular, and functional genomic techniques in a wide array of > taxa to interrogate the evolution and development of novel traits. The > primary model organism in the lab is the sea anemone Nematostella > vectensis and we have recently developed cultures of corals and tube > anemones for comparative studies. We are a highly collaborative and > interdisciplinary group, and we aim to create a diverse and welcoming > environment that supports curiosity-driven science and exploratory > studies to maximize opportunities for discovery. For more information > on our interests, please visit: www.babonislab.com. > > Requirements > > To qualify, applicants must have a Ph.D. and expertise in molecular > biology, genetics, biochemistry, and embryonic development. The ideal > candidate will also have significant experience with functional genomics, > genome editing (including CRISPR/Cas9 technology), gene delivery > techniques (e.g. microinjection, electroporation), and DNA and RNA > sequencing and analysis. > > The position will be appointed initially for 1 year with the option > to renew pending satisfactory performance. Salary is commensurate with > NIH standards. > > To apply: > > Please apply via Academic Jobs Online > (https://ift.tt/ktyeDjA). > > Please submit a current curriculum vitae, a short cover letter describing > your current research and your specific experience with the required > and preferred techniques, a 1-page description of your future research > interests indicating how they align with the research goals of the > Babonis Lab, contact information for two professional references, and > a statement of contribution to diversity, equity and inclusion via the > website. Review of applications will begin April 1, 2023. The preferred > start date for the position is June 1, 2023. > > Applications will be reviewed as received, continuing until a suitable > applicant is identified. > > Salary range is $56,484 -$75,000 > > Cornell University is an innovative Ivy League university and a great > place to work. Our inclusive community of scholars, students, and staff > impart an uncommon sense of larger purpose and contribute creative > ideas to further the university's mission of teaching, discovery, > and engagement. > > Diversity and Inclusion are a part of Cornell University’s heritage. We > are a recognized employer and educator valuing AA/EEO, Protected Veterans, > and Individuals with Disabilities. > > We also recognize a lawful preference in employment practices for Native > Americans living on or near Indian reservations. > > > Leslie Babonis
0 notes
Text
Organoids: from traditional 2D cell Culture to 3D culture models
In recent years, organoid culture technology is developing rapidly. Organoids has been awarded 'Method of the Year 2017' by Nature Methods, for their immense potential as tools to study human biology in health and disease.
Previously, the term ‘organoid’ has been used to encompass all 3D organotypic cultures derived from primary tissues, pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), established cell lines, as well as whole or segmented organs such as organ explants consisting of multiple tissue types. The ‘organoid’ was defined by Fatehullah et al. As an in vitro 3D cellular cluster derived exclusively from primary tissue, ESCs or iPSCs, capable of self-renewal and self-organization, and exhibiting similar organ function as the tissue of origin[1-3].
The development of organoids
Organoid technology is built upon the foundation of stem cell technologies, classical developmental biology and cell-mixing experiments. In the early 20th century, Wilson (1907) demonstrated that dissociated sponge cells can self-organize to regenerate a whole organism. Stem cell research began to thrive when murine ESCs (mESCs) were first isolated and established in 1981[4-6]. In 2009, Hans Clevers lab established a long-term primary culture to generate the intestinal organoid culture system. It was an outstanding technological leap for the stem cell field[5][7].
Lately, organoids have been successfully generated for an increasing variety of organs, including but not limited to gut, stomach, lung, kidney, liver, pancreas, mammary glands, prostate, thyroid, retina, inner ear, taste bud and brain[8].
Organoid model is a major technological breakthrough that acts as a valuable model for the study of tissue development, disease modeling, drug screening, personalized medicine and cell therapy[1].、
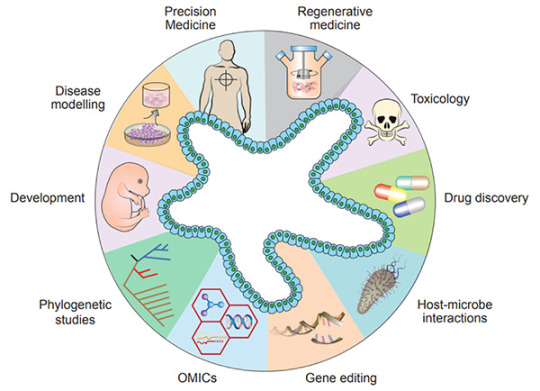
Figure 1. Diverse applications of organoid technology
[5]
.
Recently, organoids have been widely used in many areas, including developmental biology, disease modeling, precision medicine, regenerative medicine, toxicology, drug discovery studies, host-microbiome interactions, gene editing, multi-omics, and phylogenetic studies[5].
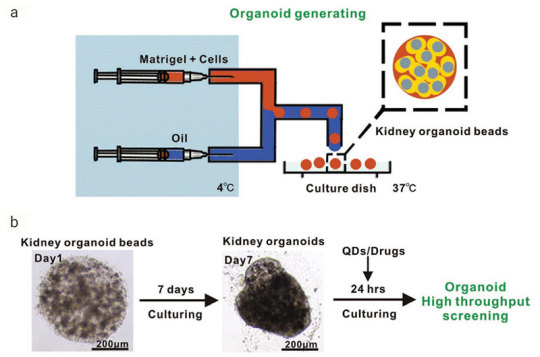
Figure 2. Generation of Reproducible Kidney Organoids
[9]
.Mouse kidney cells are suspended in Matrigel liquid precursor and fabricated organoid beads by microfluid machine and 3D printer. Organoid beads are cultured supplement with
Noggin
,
R-spondin 1
,
FGF-4
,
FGF-basic
,
SB-431542
,
CHIR-99021
. The size, shape, and composition of the kidney organoids are highly reproducible.Advantages of organoid model
Organoids represent an important bridge between 2D cultures and in vivo mouse/human models. They are more physiologically relevant than monolayer culture models and are far more amenable to manipulation of niche components, signaling pathways and genome editing than in vivo models [1][10]. Some of the advantages of the organoid models are;
1) Compared to traditional two-dimensional (2D) cell culture, organoids are similar to primary tissue in both their composition and architecture, harboring small populations of genomically stable, self-renewing stem cells that give rise to fully differentiated progeny comprising all major cell lineages at frequencies similar to those in living tissues[11].
2) Organoids can be expanded enormously, cryopreserved as biobanks, and easily manipulated using techniques similar to those established for 2D monolayer culture[11].
3) Primary-tissue-derived organoids lack mesenchyme/stroma that provides a separate system for studying a specific tissue of interest without being influenced by the local microenvironment[1].
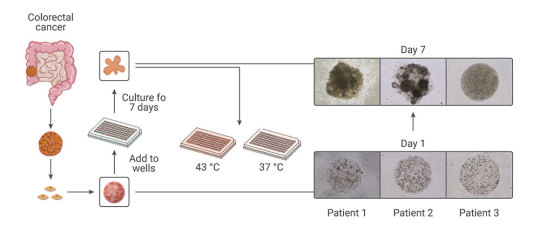
Figure 3. Comparison of Organoid Cultures with Two-Dimensional Cell Cultures and Studies in Animals
[10]
.
Compared with the traditional patient-derived cancer cell line (PDC) and patient-derived xenograft (PDX) model, the PDO model has unparalleled advantages. In the screening of drugs for tumor therapy, tumor organoid models derived from patient tumors have higher sensitivity, heterogeneity, and stability and can restore the genuine attributes of tumors more effectively. In addition, tumor organoids can be preserved, resuscitated, passed infinitely, and mechanically cultured on a chip for drug screening[13]. Therefore, organoid technology exerts enormous potential in evaluation of efficacy and toxicity of drugs, regenerative medicine, and precision medicine. Organoids have been established successfully for multiple cancer types, including but not limited to stomach cancer, colorectal cancer, liver cancer, pancreatic cancer[14].
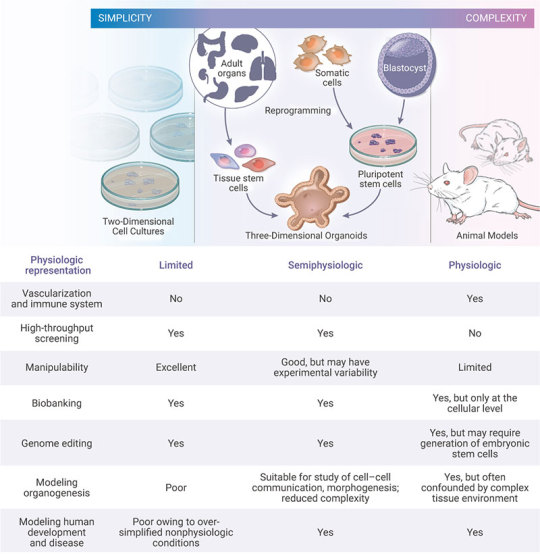
Figure 4. Establishment of patient-derived organoids as in vitro tumor models for colorectal cancer
[15]
.
Many pathogenic viruses that infect humans display species specificity and animal models can’t be used to study those viral infections. Hence, studying viral biology and identifying potential treatments benefits by developing in vitro cell systems (organoids) that closely mimic human physiology. In the current COVID-19 pandemic, organoids have emerged as powerful tools for SARS-CoV-2 research, bridging the gap between cell lines and in vivo animal models[16-17].
The "Magic" in organoid culture medium
Organoids can be generated from tissue-resident adult stem cells (ASCs) or from PSCs. Under appropriate conditions, supplementation of proper culture medium, growth factors and small molecules, stem cells embedded in Matrigel can undergo continuous self-renewal and differentiation, and self-organize into 3D-structures. The methods of culturing different organoids are similar[17].
1) Multiple sources:
1.1). ASCs-derived organoids: Primary tissue that is dissociated into functional sub-tissue units containing stem cells. These functional units are further digested into single cells and FACS-sorted to enrich for stem cells.
1.2). ESCs/iPSCs-derived organoids: stem cells undergo directed differentiation towards the desired germ lineage, eventually generating floating spheroids that are subsequently embedded in extracellular matrix (ECM) to initiate organoid culture[1].
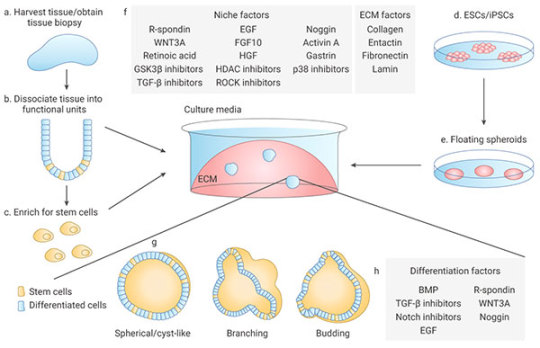
Figure 5. Organoid generation and culture from primary tissue and ESCs/iPSCs
[2]
.
2) Manipulability of niche components:
Organoids are typically cultured in an ECM surrounded by culture media supplemented with specific niche factors (different from air-liquid interface (ALI) method which is introduced recently)[18]. Organoids can either differentiate spontaneously or be induced to differentiate towards desired lineages or cell types by adding suitable differentiation factors and/or withdrawing factors that promote stemness. Common niche and ECM factors include R-spondin, EGF, Noggin, Activin A, and Collagen. Specific small molecules are added such as TGF-β inhibitor A-83-01, GSK3β inhibitor CHIR99021, and ROCK inhibitor Y27632[1].
Stem cells are maintained and perpetuated in organoids, continually giving rise to differentiated progeny. In addition, organoids can be dissociated and plated onto membrane supports coated with Matrigel or Collagen to form 2D monolayer organoid models[17].
MedChemExpress offers a variety of high-quality recombinant proteins and small molecules for organoid culture.
Related products
Cytokines
Human EGF
A well-known growth factor for epithelial tissues; binding to EGF receptors, induces hyperplasic changes.
EGF can be used for the generation of Gastrointestinal tract, liver, thyroid, brain organoids.
Human FGF-2/4/9/10
FGFs play crucial roles in a wide variety of cellular functions, including cell proliferation, survival, metabolism, morphogenesis, and differentiation,
as well as in tissue repair and regeneration. In a 3D extracellular matrix, FGF-2, FGF-7, FGF-9, and FGF-10 promote lung organoid formation.
Human HGF
HGF is a known hepatocyte mitogen that can be used for the liver organoid culture.
Human Wnt3a
Wnt is a master regulator in regulation of cell development, proliferation, differentiation, adhesion, and polarity.
Wnt3a is an essential niche component for maintaining the proliferation of Lgr5-positive stem cells in various organoids such as the small intestine,
large intestine, stomach, pancreas and liver.
Human BMP-4
BMPs play crucial roles in embryogenesis and development, and also in maintenance of adult tissue homeostasis.
BMP-2 and BMP-4 are widely used in in vitro protocols of generation of hepatic cells from induced pluripotent stem cells (iPS) and from embryonic stem cells (ESC).
Human Noggin
Noggin is an inhibitor of bone morphogenetic proteins that modulates cellular differentiation, proliferation, and apoptosis.
Noggin is one of the most important components of organoid media are growth factors.
Human DKK-1
DKK-1 is a canonical WNT inhibitor that can induce retinal progenitors to self-organize.
Small-molecule Inhibitor
Y-27632 dihydrochloride
Y-27632 is a Rho Kinase (ROCK) inhibitor; Has been used to increase the proliferation and reduce apoptosis of progenitor cells grown in vitro.
A 83-01
A 83-01 is an inhibitor of TGF-β type I receptor ALK5, the Activin/Nodal receptor ALK4, and the nodal receptor ALK7.
References:
[1] Aliya Fatehullah, Si Hui Tan, Nick Barker, et al. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016 Mar;18(3):246-54.
[2] Marina Simian, Mina J Bissell. Organoids: A historical perspective of thinking in three dimensions. J Cell Biol. 2017 Jan 2;216(1):31-40.
[3] HansClevers. Modeling Development and Disease with Organoids. Cell. 2016 Jun 16;165(7):1586-1597.
[4] Madeline A Lancaster, Juergen A. Knoblich. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014 Jul 18;345(6194):1247125.
[5] Claudia Corrò, Vivian S.W. Li, et al. A brief history of organoids. Am J Physiol Cell Physiol. 2020 Jul 1;319(1):C151-C165.
[6] G R Martin. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7634-8.
[7] Sato T, Vries RG, et al. (2009). “Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche.” Nature 459(7244): 262–265.
[8] Elisa Suarez Martinez , Amancio Carnero, et al. 3D and organoid culture in research: physiology, hereditary genetic diseases and cancer. Cell Biosci. 2022; 12: 39.
[9] Chengyong He, Shaohua Ma, Zhenghong Zuo, et al. Black Phosphorus Quantum Dots Cause Nephrotoxicity in Organoids, Mice, and Human Cells. Small. 2020 Jun;16(22):e2001371.
[10] Mo Li, Juan C Izpisua Belmonte. Organoids-Preclinical Models of Human Disease. N Engl J Med. 2019 Feb 7;380(6):569-579.
[11] Mariangela Scalise, Fabiola Marino, Daniele Torella, et al. From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish. Int J Mol Sci. 2021 Dec; 22(24): 13180.
[12] Xialin Nie, Zhixing Liang, Linsen Ye, Yang Yang, et al. Novel organoid model in drug screening: Past, present, and future. Liver Research 5 (2021) 72-78.
[13] Chen Liu , Chaoyang Sun , et al. Drug screening model meets cancer organoid technology. Transl Oncol. 2020 Nov; 13(11): 100840.
[14] Hanxiao Xu, Kongming Wu, et al. Organoid technology and applications in cancer research. J Hematol Oncol 11, 116 (2018).
[15] Lisi Zeng, Shuzhong Cui, Shengwei Jiang, et al. Raltitrexed as a synergistic hyperthermia chemotherapy drug screened in patient-derived colorectal cancer organoids. Cancer Biol Med. 2021 Mar 12;18(3):750-762.
[16] Maarten H.Geurts, Jeltevan der Vaart, HansClevers, et al. The Organoid Platform: Promises and Challenges as Tools in the Fight against COVID-19. Volume 16, Issue 3, 9 March 2021, Pages 412-418.
[17] Jelte van der Vaart, Mart M. Lamers, Hans Clevers, et al. Advancing lung organoids for COVID-19 research. Dis Model Mech. 2021 Jun 1; 14(6): dmm049060.
[18] Soumya K Kar, et al. Organoids: a promising new in vitro platform in livestock and veterinary research. Vet Res. 2021 Mar 10;52(1):43.
[19] Yaqi Li, Guoqiang Hua, et al. Organoid based personalized medicine: from bench to bedside. Cell Regen. 2020 Dec; 9: 21.
0 notes
Text
Researchers at Columbia create 40 people, kill them after gene editing mistake
https://www.wsj.com/articles/crispr-gene-editing-can-lead-to-big-mistakes-in-human-embryos-11603983608
“...Researchers in the new study published in Cell said they used sperm from a single human donor to create 40 embryos. The man has hereditary blindness caused by a mutation on the EYS gene, which is located on chromosome 6, one of the 23 pairs in humans. The scientists used Crispr-Cas9 to cut the father’s DNA at the site of the gene mutation they wanted to correct. When a cut is made in the DNA, the cell tries to repair the DNA.
“Scientists injected a Crispr-Cas9 enzyme into 37 embryos, and three other embryos served as controls. The scientists found that about half of the embryos lost large segments of the chromosome, or the entire chromosome, on which the EYS gene is located. “This is a very adverse outcome,” Dr. Egli said.”
[Full article below the break.]
Crispr Gene Editing Can Lead to Big Mistakes in Human Embryos
Columbia University study of Crispr technology found it made unwanted chromosomal changes in human embryos
Scientists using the Crispr gene-editing technology in human embryos to try to repair a gene that causes hereditary blindness found it made unintended and unwanted changes, frequently eliminating an entire chromosome or large sections of it.
The study published Thursday in the journal Cell comes as the international scientific community continues to grapple with the potential use of Crispr for editing human embryos that would be intended for creating a pregnancy and birth.
In September, an international commission sponsored by the U.S. National Academy of Medicine, U.S. National Academy of Sciences and the U.K.’s Royal Society issued a report stating that the gene-editing technology isn’t ready for such a use because scientists don’t understand how to make precise fixes without also introducing potentially dangerous changes.
Dieter Egli, assistant professor of developmental cell biology at Columbia University and the study’s senior author, said, “This study is not going to stop the field. But we have to ask what to do with these powerful tools, and in which context they are safe and efficacious.”...
Two separate papers published earlier this month indicate that the ethical debate continues over whether and under what circumstances creating genetically modified children could be permissible.
One paper, published in the Crispr Journal, solicited views of more than three dozen experts on issues raised in the international commission’s September report on Crispr germ-line editing. The technique—which involves making changes to eggs, sperm and embryos—is controversial because any changes can be passed to future generations. The paper revealed major differences of opinion among leading scholars, including Jennifer Doudna, the University of California, Berkeley, biochemist who with a colleague was awarded the Nobel Prize in chemistry this year for pioneering work on Crispr.
In her comments on the report, Dr. Doudna said the commission’s recommendations reflect consensus in the field that the technology shouldn’t be used for embryo editing in the clinic at this time. She added that she was struck by the inclusion of certain diseases that are already being managed, such as cystic fibrosis, with disorders where embryo editing might someday be permissible.
The other paper, also published in the Crispr Journal, surveyed policies across 106 countries regarding germ-line gene editing. The researchers found 96 of the countries already had applicable policy documents, such as legislation, regulations or international treaties. Of those 96 countries, 75 bar the use of genetically modified embryos for the purpose of starting a pregnancy—an indication that it might be possible to create an international consensus on the issue, the researchers said.
Crispr enables scientists to cut, edit and insert DNA and has been an object of excitement, fascination and controversy since its discovery in 2012. The technology opens up the possibility of treating severe illness for which there are no effective therapies. Trials are under way or expected to start soon in patients with cancer, sickle-cell anemia and other conditions in China, Europe and the U.S.
The potential to use Crispr to create genetically modified babies has been an area of major concern, especially after the 2018 announcement of the birth of twin girls from embryos whose DNA had been changed using the Crispr technology.
A second woman implanted with a genetically modified embryo is also believed to have given birth.
Very little is known about the health of the babies. He Jiankiu, the Chinese researcher who spearheaded the experiments, was sentenced to three years in prison after being found guilty last year in a court in China of conducting illegal medical practices.
Researchers in the new study published in Cell said they used sperm from a single human donor to create 40 embryos. The man has hereditary blindness caused by a mutation on the EYS gene, which is located on chromosome 6, one of the 23 pairs in humans.
The scientists used Crispr-Cas9 to cut the father’s DNA at the site of the gene mutation they wanted to correct. When a cut is made in the DNA, the cell tries to repair the DNA.
Scientists injected a Crispr-Cas9 enzyme into 37 embryos, and three other embryos served as controls.
The scientists found that about half of the embryos lost large segments of the chromosome, or the entire chromosome, on which the EYS gene is located.
“This is a very adverse outcome,” Dr. Egli said.
Scientists still don’t know a lot about the mechanisms of human embryo development. The U.S. government doesn’t permit the use of federal funding to conduct research on human embryos. The research for the Cell study was funded by private donations from the New York Stem Cell Foundation and the Russell Berrie Foundation Program in Cellular Therapies.
Dr. Egli said scientists know more about the cell and molecular biology of flies, worms and mice embryos than human embryos. He said the current study seems to indicate that “the human embryo seems unique in its poor ability to fix a DNA break.” The significance of the biological finding “goes beyond the debate” over the use of Crispr in embryos, he said.
Nonetheless, the ethical debate will likely continue alongside development of the technology. Françoise Baylis, university research professor at Dalhousie University in Canada, one of the authors on both Crispr Journal papers, said that much of the debate within the scientific community over editing human embryos focused on whether the technology is safe and effective enough to use. “That’s not the right question to start with,” Dr. Baylis said. “I am not asking if the science is ready. I am asking what problems do we want to solve, and what science will help us solve them.” Dr. Baylis is a member of a World Health Organization expert advisory panel set up last year to issue global standards and guidelines for editing of the human genome. The panel hasn’t yet issued its guidance.
Marcy Darnovsky of the Center for Genetics and Society in Berkeley, Calif., a public-interest advocacy group in human genetics and assisted reproductive technologies and one of the authors of the global policy study, said the Cell paper indicates the substantial technical challenges that remain surrounding Crispr gene editing of embryos. “It gives us time,” she said. “There is still a window to have a meaningful conversation about the social questions.”
9 notes
·
View notes
Note
So terf that can’t read you gonna give actual scientific evidence or not. Let’s see if you answer this or do you like hiding in the notes.
lol ok. Here’s medical journals and textbook citations to keep you up at night
“Overall, 95% of all biologists affirmed the biological view that a human's life begins at fertilization (5212 out of 5502). “
https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3211703
"The development of a human being begins with fertilization, a process by which two highly specialized cells, the spermatozoon from the male and the oocyte from the female, unite to give rise to a new organism, the zygote."
[Langman, Jan. Medical Embryology. 3rd edition. Baltimore: Williams and Wilkins, 1975, p. 3]
"Embryo: An organism in the earliest stage of development; in a man, from the time of conception to the end of the second month in the uterus."
[Dox, Ida G. et al. The Harper Collins Illustrated Medical Dictionary. New York: Harper Perennial, 1993, p. 146]
"Almost all higher animals start their lives from a single cell, the fertilized ovum (zygote)... The time of fertilization represents the starting point in the life history, or ontogeny, of the individual."
[Carlson, Bruce M. Patten's Foundations of Embryology. 6th edition. New York: McGraw-Hill, 1996, p. 3]
http://www.lifeissues.net/writers/imb/imb_01lifebegins.html
"Human development begins after the union of male and female gametes or germ cells during a process known as fertilization (conception).
"Fertilization is a sequence of events that begins with the contact of a sperm (spermatozoon) with a secondary oocyte (ovum) and ends with the fusion of their pronuclei (the haploid nuclei of the sperm and ovum) and the mingling of their chromosomes to form a new cell. This fertilized ovum, known as a zygote, is a large diploid cell that is the beginning, or primordium, of a human being."
[Moore, Keith L. Essentials of Human Embryology. Toronto: B.C. Decker Inc, 1988, p.2]
The chromosomes of the oocyte and sperm are...respectively enclosed within female and male pronuclei. These pronuclei fuse with each other to produce the single, diploid, 2N nucleus of the fertilized zygote. This moment of zygote formation may be taken as the beginning or zero time point of embryonic development."
[Larsen, William J. Human Embryology. 2nd edition. New York: Churchill Livingstone, 1997, p. 17]
"Although life is a continuous process, fertilization is a critical landmark because, under ordinary circumstances, a new, genetically distinct human organism is thereby formed.... The combination of 23 chromosomes present in each pronucleus results in 46 chromosomes in the zygote. Thus the diploid number is restored and the embryonic genome is formed. The embryo now exists as a genetic unity."
[O'Rahilly, Ronan and M�ller, Fabiola. Human Embryology & Teratology. 2nd edition. New York: Wiley-Liss, 1996, pp. 8, 29. This textbook lists "pre-embryo" among "discarded and replaced terms" in modern embryology, describing it as "ill-defined and inaccurate" (p. 12}]
#immensely satisfied to be arguing so well I’m living in your head rent free. transphobes mad#the funny thing is I used to be more pro choice#and then I listened to the pro choice people and they’re all eugenecists#politics#my post
54 notes
·
View notes
Photo

Panel Lays Out Guidelines for CRISPR-Edited Human Embryos
By Lisa Winter (The-Scientist). Image: © ISTOCK.COM, POSTERIORI
On September 3, 2020, the International Commission on the Clinical Use of Human Germline Genome Editing released a report that reviews the available research and determines gene editing’s ethical use on human embryos. The 225-page document offers a roadmap to the testing and regulations necessary to develop the technology and ultimately concludes that the technology is not yet reliable enough to use on humans. Any country that permits its scientists to do so in the future should limit the activity to severe single-gene diseases.
The commission, composed of dozens of scientists worldwide, was formed after Chinese geneticist He Jiankui claimed in 2018 to have used CRISPR-Cas9 on a set of twins and a third baby to make them HIV-resistant. While the validity of the editing has yet to be determined, He’s claims have been met with international condemnation and a three-year prison sentence for illegally practicing medicine.
Somatic gene editing introduces altered DNA into some of the body’s cells post-development as a means to treat genetic conditions. Germline editing manipulates DNA in the earliest stages of embryonic development, affects all tissue types, and permits the organism to then pass down those alterations to their offspring. Instead of treating the disease, germline editing would theoretically be able to eliminate it not only from the organism but from its lineage completely, making it attractive in the context of heritable genetic diseases.
Although manipulating genomes via CRISPR has become commonplace in genetic labs around the world, the commission states that, currently, “the outcomes of genome editing in human zygotes cannot be adequately controlled” and could bear unintended consequences.
“It underscores what really I think most researchers who think about this are aware of: There must not be any use of germline editing for clinical purposes at this time,” Jennifer Doudna, a geneticist at the University of California, Berkeley, who was not a member of the commission, tells STAT. “And the reason is the technology is just too early-stage and we don’t understand well enough how it works in human embryos.”
Once it is possible to safely make these changes, the commission advises, countries could adopt policies allowing this technology only for the cases when a single gene is responsible for the disorder, such as sickle cell anemia, cystic fibrosis, or Tay-Sachs.
“We think the bar should be high, and appropriately so,” Richard Lifton, cochair of the panel, tells The Guardian. “If you are going to be creating human beings, you want to know that you can reliably make the edits you’re intending. If you can’t do it reliably, without introducing unintended effects, you shouldn’t be going [forward].”
53 notes
·
View notes
Text
Living robots built using frog cells
A book is made of wood. But it is not a tree. The dead cells have been repurposed to serve another need.

Now a team of scientists has repurposed living cells -- scraped from frog embryos -- and assembled them into entirely new life-forms. These millimeter-wide "xenobots" can move toward a target, perhaps pick up a payload (like a medicine that needs to be carried to a specific place inside a patient) -- and heal themselves after being cut.
"These are novel living machines," says Joshua Bongard, a computer scientist and robotics expert at the University of Vermont who co-led the new research. "They're neither a traditional robot nor a known species of animal. It's a new class of artifact: a living, programmable organism."
The new creatures were designed on a supercomputer at UVM -- and then assembled and tested by biologists at Tufts University. "We can imagine many useful applications of these living robots that other machines can't do," says co-leader Michael Levin who directs the Center for Regenerative and Developmental Biology at Tufts, "like searching out nasty compounds or radioactive contamination, gathering microplastic in the oceans, traveling in arteries to scrape out plaque."
The results of the new research were published January 13 in the Proceedings of the National Academy of Sciences.
youtube
Bespoke Living Systems
People have been manipulating organisms for human benefit since at least the dawn of agriculture, genetic editing is becoming widespread, and a few artificial organisms have been manually assembled in the past few years -- copying the body forms of known animals.
But this research, for the first time ever, "designs completely biological machines from the ground up," the team writes in their new study.
With months of processing time on the Deep Green supercomputer cluster at UVM's Vermont Advanced Computing Core, the team -- including lead author and doctoral student Sam Kriegman -- used an evolutionary algorithm to create thousands of candidate designs for the new life-forms. Attempting to achieve a task assigned by the scientists -- like locomotion in one direction -- the computer would, over and over, reassemble a few hundred simulated cells into myriad forms and body shapes. As the programs ran -- driven by basic rules about the biophysics of what single frog skin and cardiac cells can do -- the more successful simulated organisms were kept and refined, while failed designs were tossed out. After a hundred independent runs of the algorithm, the most promising designs were selected for testing.
Then the team at Tufts, led by Levin and with key work by microsurgeon Douglas Blackiston -- transferred the in silico designs into life. First they gathered stem cells, harvested from the embryos of African frogs, the species Xenopus laevis. (Hence the name "xenobots.") These were separated into single cells and left to incubate. Then, using tiny forceps and an even tinier electrode, the cells were cut and joined under a microscope into a close approximation of the designs specified by the computer.
Assembled into body forms never seen in nature, the cells began to work together. The skin cells formed a more passive architecture, while the once-random contractions of heart muscle cells were put to work creating ordered forward motion as guided by the computer's design, and aided by spontaneous self-organizing patterns -- allowing the robots to move on their own.
These reconfigurable organisms were shown to be able move in a coherent fashion -- and explore their watery environment for days or weeks, powered by embryonic energy stores. Turned over, however, they failed, like beetles flipped on their backs.
Later tests showed that groups of xenobots would move around in circles, pushing pellets into a central location -- spontaneously and collectively. Others were built with a hole through the center to reduce drag. In simulated versions of these, the scientists were able to repurpose this hole as a pouch to successfully carry an object. "It's a step toward using computer-designed organisms for intelligent drug delivery," says Bongard, a professor in UVM's Department of Computer Science and Complex Systems Center.
Living Technologies
Many technologies are made of steel, concrete or plastic. That can make them strong or flexible. But they also can create ecological and human health problems, like the growing scourge of plastic pollution in the oceans and the toxicity of many synthetic materials and electronics. "The downside of living tissue is that it's weak and it degrades," say Bongard. "That's why we use steel. But organisms have 4.5 billion years of practice at regenerating themselves and going on for decades." And when they stop working -- death -- they usually fall apart harmlessly. "These xenobots are fully biodegradable," say Bongard, "when they're done with their job after seven days, they're just dead skin cells."
Your laptop is a powerful technology. But try cutting it in half. Doesn't work so well. In the new experiments, the scientists cut the xenobots and watched what happened. "We sliced the robot almost in half and it stitches itself back up and keeps going," says Bongard. "And this is something you can't do with typical machines."
Cracking the Code
Both Levin and Bongard say the potential of what they've been learning about how cells communicate and connect extends deep into both computational science and our understanding of life. "The big question in biology is to understand the algorithms that determine form and function," says Levin. "The genome encodes proteins, but transformative applications await our discovery of how that hardware enables cells to cooperate toward making functional anatomies under very different conditions."
To make an organism develop and function, there is a lot of information sharing and cooperation -- organic computation -- going on in and between cells all the time, not just within neurons. These emergent and geometric properties are shaped by bioelectric, biochemical, and biomechanical processes, "that run on DNA-specified hardware," Levin says, "and these processes are reconfigurable, enabling novel living forms."
The scientists see the work presented in their new PNAS study -- "A scalable pipeline for designing reconfigurable organisms," -- as one step in applying insights about this bioelectric code to both biology and computer science. "What actually determines the anatomy towards which cells cooperate?" Levin asks. "You look at the cells we've been building our xenobots with, and, genomically, they're frogs. It's 100% frog DNA -- but these are not frogs. Then you ask, well, what else are these cells capable of building?"
"As we've shown, these frog cells can be coaxed to make interesting living forms that are completely different from what their default anatomy would be," says Levin. He and the other scientists in the UVM and Tufts team -- with support from DARPA's Lifelong Learning Machines program and the National Science Foundation -- believe that building the xenobots is a small step toward cracking what he calls the "morphogenetic code," providing a deeper view of the overall way organisms are organized -- and how they compute and store information based on their histories and environment.
Future Shocks
Many people worry about the implications of rapid technological change and complex biological manipulations. "That fear is not unreasonable," Levin says. "When we start to mess around with complex systems that we don't understand, we're going to get unintended consequences." A lot of complex systems, like an ant colony, begin with a simple unit -- an ant -- from which it would be impossible to predict the shape of their colony or how they can build bridges over water with their interlinked bodies.
"If humanity is going to survive into the future, we need to better understand how complex properties, somehow, emerge from simple rules," says Levin. Much of science is focused on "controlling the low-level rules. We also need to understand the high-level rules," he says. "If you wanted an anthill with two chimneys instead of one, how do you modify the ants? We'd have no idea."
"I think it's an absolute necessity for society going forward to get a better handle on systems where the outcome is very complex," Levin says. "A first step towards doing that is to explore: how do living systems decide what an overall behavior should be and how do we manipulate the pieces to get the behaviors we want?"
In other words, "this study is a direct contribution to getting a handle on what people are afraid of, which is unintended consequences," Levin says -- whether in the rapid arrival of self-driving cars, changing gene drives to wipe out whole lineages of viruses, or the many other complex and autonomous systems that will increasingly shape the human experience.
497 notes
·
View notes
Link
0 notes
Text
“As the biologist on this panel of philosophers, I thought I’d leave them to the tricky job of discussing the ethical concerns of designer babies, and instead focus on the practical aspects. Gene editing is a far more difficult endeavour than you might imagine, because many people have a very naive idea of how genetics, and the relationship between genotype and phenotype, works. So I’ll just bring up a couple of biological concerns that would mean I’d never consider having a gene-edited child.
Very few genes act alone; they’re part of a network of other genes that work together to assemble the organism. How big and complex is that network? According to the omnigenic model, every gene influences and is influenced by every other gene, which means the background genetics of 20,000 genes has to be considered when you manipulate any one gene. You might wonder, then, how this could work for all those research scientists who are busily doing experimental analyses of genes edited in their lab mice, or fruit flies. The answer: lab research animals are highly inbred, with relatively little genetic variation between individuals. That means you can tinker with the genetic makeup of one individual, and get replicable results in another animal of the same strain. Humans are much messier than that. You should know this from all the pharmaceutical ads thrown at us: there are all kinds of side effects and unpredictable variations in medications. One pill might work great for you, be terrible for your friend at work, and do nothing at all for your Uncle Ralph. Now imagine the same range of side-effects that you might inflict on a baby, and realise that there’s no way to “stop taking” the gene editing pill, and they will have to live with it their entire life.
What gene modification do you want? Because we honestly don’t know what most genes do, at least at the level of knowing all of their effects. There are a few disease genes of large effect we can dream of eradicating, such as the genes for phenylketonuria, or sickle cell anaemia, or cystic fibrosis. Those are problems so severe that the choice is between a certainty of a short, sick life, or the risks of experimental gene modification.
There is a lot of misplaced confidence in genetic engineering. Sure, we can edit out a defective gene in a mouse embryo — with some sloppiness. Sometimes the wrong bit of DNA is spliced out. Sometimes the insertion leaves little molecular scars around the site. Sometimes it fails in some of the embryonic cells, so some have the modification and others don’t. Sometimes the scientist doesn’t actually know what they’re doing.
One other significant concern is the motivation behind gene-editing embryos, and here I’m segueing into the moral concerns, which I’ll leave to the philosophers. Beyond editing out known, serious single gene defects, why do people want this? Too often we’re drifting neatly into eugenics, in which certain properties of people are valued more than certain others, but rather than executing the unfit, we’re going to selectively promote other people with properties we consider “better”. For instance, there are people who want to make sure none of their children are gay, so if we could zap the gay gene (which doesn’t exist, by the way), we could make sure that everyone is suitably heterosexual. To which I have to say three things:
But what about, for example, if someone wants their son to be destined to become a world-class football player. What genes would you want modified? It’s going to require a lot, and we don’t know what all they are. You can’t just simply switch on a big muscle gene — we do know how to do that! It’s been done in a cattle breed, the Belgian Blue, by a natural mutation in the myostatin gene, which normally keeps muscle growth under control. But there’s more to being a great athlete than having giant muscles! These cows also have serious side-effects: larger calves and smaller birth canals mean most have to be delivered by caesarian, and the adults have cardiovascular and joint issues.
Gene-editing for athleticism is worse than a gamble, because you’re guaranteed to fail to modify necessary complementary genes, you don’t know what all genes are needed for great athletic performance, you’ve just added a poorly understood variable to the poorly understood recipe for athleticism, and you’ve also added the potential for deleterious effects due to the manipulation required.
There’s already a better solution: you want a great football player, then practice, practice, practice. Raise them in an environment where football is important and they learn to love the game, and you’ve got a better chance of raising a football player than randomly mucking around in their genome. This is also true of other traits, like if you want a smart kid. We already know how to maximise that — it’s called an education.
For example, the Chinese scientist He Jiankui thought he’d engineer HIV resistance into the embryos of parents with HIV. Sounds like a good idea, right? Except that his idea was to mimic a known natural mutation called CCR-delta-32, which is carried in some people, especially Northern Europeans, who have reduced incidence of HIV in homozygous carriers.
The problem: it involves deleting some of the immune system machinery, which HIV uses to recognise immune system cells. We don’t know what else about the immune system is affected by the change. The resistance requires that both copies of the CCR-delta-32 gene be removed, but He’s changes only knocked out one copy, so it would be effectively ineffectual. The children modified by his procedure were also mosaic, so only some of their cells were affected.
The only thing his experiment accomplished was to make him notorious. And put him in prison, apparently.
What’s wrong with being gay?
What if deleting gayness also deleted other properties of the person?
And hey, you do know nobody knows how to do that, even if they had a flawless CRISPR/Cas9 protocol, right?”
- PZ Myers
#bioethics#academia#pz myers#long post#not really relevant to this blog#but very interesting and important
12 notes
·
View notes
Text
Fwd: Postdoc: USDA_GainsvilleFlorida.SexRatioEvolution
Begin forwarded message: > From: [email protected] > Subject: Postdoc: USDA_GainsvilleFlorida.SexRatioEvolution > Date: 14 December 2022 at 11:05:58 GMT > To: [email protected] > > > Post-doctoral Research Associate Position > Transgenic approaches to sex ratio distortion and inducing male sterility > USDA-ARS Center for Medical, Agricultural, and Veterinary Entomology > > A post-doctoral position working with Al Handler at the USDA-ARS-CMAVE > lab in Gainesville, FL is available immediately. The goals of > this postdoc will be to create genetically modified tephritid and > drosophilid fruit fly strains for conditional sex-specific selection > and male sterility. Ultimately, these strains may become tools for > population control programs. There will also be opportunities to study > the genetic breakdown of transgenic strains, a growing concern given the > massive development of transgenic approaches to population control. These > projects are funded by the USDA-NIFA Biotechnology Risk Assessment Program > and contribute to ongoing collaborative FAO/IAEA Coordinated Research > Projects. Furthermore, this postdoc is a collaborative effort with Dan > Hahn's lab at the Entomology and Nematology Dept., University of Florida > and will be administered through the University of Florida. Funding is > initially available for two-years with the possibility of extension. See > the end of this advertisement for some recent representative publications > from our group related to these topics. > > Required qualifications: The successful candidate will be expected > to contribute to ongoing research while developing an independent > research program having common objectives and will have a Ph.D. degree > and experience in one or more fields of molecular biology, genetics, > genomics and/or entomology. > > Preferred qualifications: Theoretical knowledge and bench experience > in insect molecular biological methodologies that include CRISPR-Cas9 > gene-editing, eucaryotic gene-transfer (e.g. germ-line transformation), > RNAi, molecular cloning, PCR and RT-qPCR. > > Salary: Commensurate with qualifications, experience and UF guidelines > Location: > Handler Laboratory > Insect Behavior and Biocontrol Research Unit > USDA-ARS Center for Medical, Agricultural, and Veterinary Entomology > 1700 SW 23rd Drive, Gainesville, Florida > Contact: Contact or send CV describing research background and references > to Dr. Alfred Handler: [email protected] > > Some Recent Example References: > > AM Handler, MF Schetelig, 2020. The hAT-family transposable element, > hopper, from Bactrocera dorsalis is a functional vector for insect > germline transformation. BMC genetics 21 (2), 1-9 > > GWC Thomas, E Dohmen, DST Hughes, SC Murali, M Poelchau,...AM > Handler...2020. Gene content evolution in the arthropods. Genome biology > 21 (1), 1-14 > > Y Zhao, MF Schetelig, AM Handler. 2020. Genetic breakdown of a Tet-off > conditional lethality system for insect population control. Nature > Communications 11 (1), 1-9 > > W Peng, S Yu, AM Handler, Z Tu, G Saccone, Z Xi, H Zhang. 2020. miRNA-1-3p > is an early embryonic male sex-determining factor in the Oriental fruit > fly Bactrocera dorsalis. Nature Communications 11 (1), 1-11 > > NM Teets, VS Dias, BK Pierce, MF Schetelig, AM Handler, DA > Hahn. 2019. Overexpression of an antioxidant enzyme improves male mating > performance after stress in a lek-mating fruit fly Proceedings of the > Royal Society B 286 (1904), 20190531 > > J Li, AM Handler. 2019. CRISPR/Cas9-mediated gene editing in an exogenous > transgene and an endogenous sex determination gene in the Caribbean > fruit fly, Anastrepha suspensa Gene 691, 160-166 > > > "Hahn,Daniel Allen"
0 notes
Text
Organoids: from traditional 2D cell Culture to 3D culture models
In recent years, organoid culture technology is developing rapidly. Organoids has been awarded 'Method of the Year 2017' by Nature Methods, for their immense potential as tools to study human biology in health and disease.
Previously, the term ‘organoid’ has been used to encompass all 3D organotypic cultures derived from primary tissues, pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), established cell lines, as well as whole or segmented organs such as organ explants consisting of multiple tissue types. The ‘organoid’ was defined by Fatehullah et al. As an in vitro 3D cellular cluster derived exclusively from primary tissue, ESCs or iPSCs, capable of self-renewal and self-organization, and exhibiting similar organ function as the tissue of origin[1-3].
The development of organoids
Organoid technology is built upon the foundation of stem cell technologies, classical developmental biology and cell-mixing experiments. In the early 20th century, Wilson (1907) demonstrated that dissociated sponge cells can self-organize to regenerate a whole organism. Stem cell research began to thrive when murine ESCs (mESCs) were first isolated and established in 1981[4-6]. In 2009, Hans Clevers lab established a long-term primary culture to generate the intestinal organoid culture system. It was an outstanding technological leap for the stem cell field[5][7].
Lately, organoids have been successfully generated for an increasing variety of organs, including but not limited to gut, stomach, lung, kidney, liver, pancreas, mammary glands, prostate, thyroid, retina, inner ear, taste bud and brain[8].
Organoid model is a major technological breakthrough that acts as a valuable model for the study of tissue development, disease modeling, drug screening, personalized medicine and cell therapy[1].
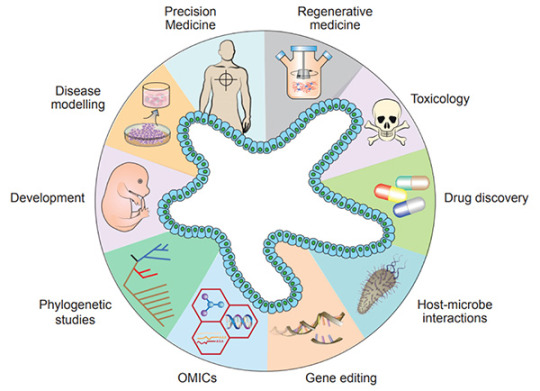
Figure 1. Diverse applications of organoid technology
[5]
.
Recently, organoids have been widely used in many areas, including developmental biology, disease modeling, precision medicine, regenerative medicine, toxicology, drug discovery studies, host-microbiome interactions, gene editing, multi-omics, and phylogenetic studies[5].
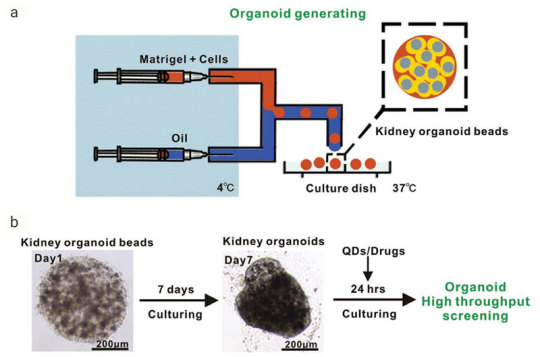
Figure 2. Generation of Reproducible Kidney Organoids
[9]
.Mouse kidney cells are suspended in Matrigel liquid precursor and fabricated organoid beads by microfluid machine and 3D printer. Organoid beads are cultured supplement with
Noggin
,
R-spondin 1
,
FGF-4
,
FGF-basic
,
SB-431542
,
CHIR-99021
. The size, shape, and composition of the kidney organoids are highly reproducible.Advantages of organoid model
Organoids represent an important bridge between 2D cultures and in vivo mouse/human models. They are more physiologically relevant than monolayer culture models and are far more amenable to manipulation of niche components, signaling pathways and genome editing than in vivo models [1][10]. Some of the advantages of the organoid models are;
1) Compared to traditional two-dimensional (2D) cell culture, organoids are similar to primary tissue in both their composition and architecture, harboring small populations of genomically stable, self-renewing stem cells that give rise to fully differentiated progeny comprising all major cell lineages at frequencies similar to those in living tissues[11].
2) Organoids can be expanded enormously, cryopreserved as biobanks, and easily manipulated using techniques similar to those established for 2D monolayer culture[11].
3) Primary-tissue-derived organoids lack mesenchyme/stroma that provides a separate system for studying a specific tissue of interest without being influenced by the local microenvironment[1].
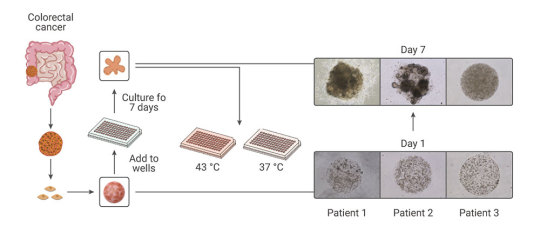
Figure 3. Comparison of Organoid Cultures with Two-Dimensional Cell Cultures and Studies in Animals
[10]
.
Compared with the traditional patient-derived cancer cell line (PDC) and patient-derived xenograft (PDX) model, the PDO model has unparalleled advantages. In the screening of drugs for tumor therapy, tumor organoid models derived from patient tumors have higher sensitivity, heterogeneity, and stability and can restore the genuine attributes of tumors more effectively. In addition, tumor organoids can be preserved, resuscitated, passed infinitely, and mechanically cultured on a chip for drug screening[13]. Therefore, organoid technology exerts enormous potential in evaluation of efficacy and toxicity of drugs, regenerative medicine, and precision medicine. Organoids have been established successfully for multiple cancer types, including but not limited to stomach cancer, colorectal cancer, liver cancer, pancreatic cancer[14].
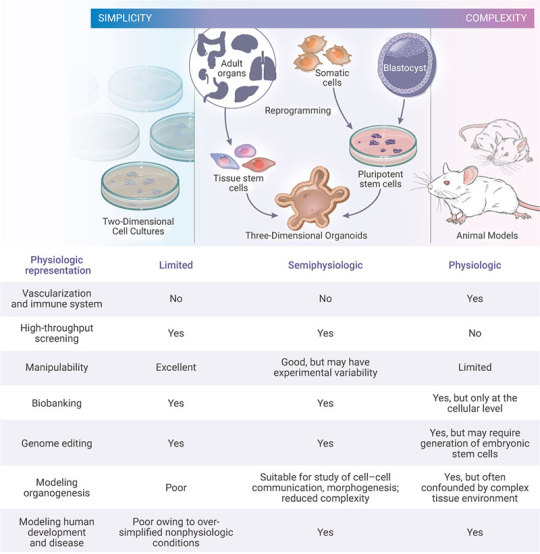
Figure 4. Establishment of patient-derived organoids as in vitro tumor models for colorectal cancer
[15]
.
Many pathogenic viruses that infect humans display species specificity and animal models can’t be used to study those viral infections. Hence, studying viral biology and identifying potential treatments benefits by developing in vitro cell systems (organoids) that closely mimic human physiology. In the current COVID-19 pandemic, organoids have emerged as powerful tools for SARS-CoV-2 research, bridging the gap between cell lines and in vivo animal models[16-17].
The "Magic" in organoid culture medium
Organoids can be generated from tissue-resident adult stem cells (ASCs) or from PSCs. Under appropriate conditions, supplementation of proper culture medium, growth factors and small molecules, stem cells embedded in Matrigel can undergo continuous self-renewal and differentiation, and self-organize into 3D-structures. The methods of culturing different organoids are similar[17].
1) Multiple sources:
1.1). ASCs-derived organoids: Primary tissue that is dissociated into functional sub-tissue units containing stem cells. These functional units are further digested into single cells and FACS-sorted to enrich for stem cells.
1.2). ESCs/iPSCs-derived organoids: stem cells undergo directed differentiation towards the desired germ lineage, eventually generating floating spheroids that are subsequently embedded in extracellular matrix (ECM) to initiate organoid culture[1].
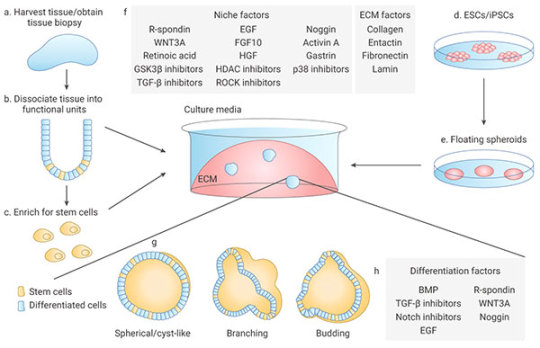
Figure 5. Organoid generation and culture from primary tissue and ESCs/iPSCs
[2]
.
2) Manipulability of niche components:
Organoids are typically cultured in an ECM surrounded by culture media supplemented with specific niche factors (different from air-liquid interface (ALI) method which is introduced recently)[18]. Organoids can either differentiate spontaneously or be induced to differentiate towards desired lineages or cell types by adding suitable differentiation factors and/or withdrawing factors that promote stemness. Common niche and ECM factors include R-spondin, EGF, Noggin, Activin A, and Collagen. Specific small molecules are added such as TGF-β inhibitor A-83-01, GSK3β inhibitor CHIR99021, and ROCK inhibitor Y27632[1].
Stem cells are maintained and perpetuated in organoids, continually giving rise to differentiated progeny. In addition, organoids can be dissociated and plated onto membrane supports coated with Matrigel or Collagen to form 2D monolayer organoid models[17].
MedChemExpress offers a variety of high-quality recombinant proteins and small molecules for organoid culture.
Related products
Cytokines
Human EGF
A well-known growth factor for epithelial tissues; binding to EGF receptors, induces hyperplasic changes.
EGF can be used for the generation of Gastrointestinal tract, liver, thyroid, brain organoids.
Human FGF-2/4/9/10
FGFs play crucial roles in a wide variety of cellular functions, including cell proliferation, survival, metabolism, morphogenesis, and differentiation,
as well as in tissue repair and regeneration. In a 3D extracellular matrix, FGF-2, FGF-7, FGF-9, and FGF-10 promote lung organoid formation.
Human HGF
HGF is a known hepatocyte mitogen that can be used for the liver organoid culture.
Human Wnt3a
Wnt is a master regulator in regulation of cell development, proliferation, differentiation, adhesion, and polarity.
Wnt3a is an essential niche component for maintaining the proliferation of Lgr5-positive stem cells in various organoids such as the small intestine,
large intestine, stomach, pancreas and liver.
Human BMP-4
BMPs play crucial roles in embryogenesis and development, and also in maintenance of adult tissue homeostasis.
BMP-2 and BMP-4 are widely used in in vitro protocols of generation of hepatic cells from induced pluripotent stem cells (iPS) and from embryonic stem cells (ESC).
Human Noggin
Noggin is an inhibitor of bone morphogenetic proteins that modulates cellular differentiation, proliferation, and apoptosis.
Noggin is one of the most important components of organoid media are growth factors.
Human DKK-1
DKK-1 is a canonical WNT inhibitor that can induce retinal progenitors to self-organize.
Small-molecule Inhibitor
Y-27632 dihydrochloride
Y-27632 is a Rho Kinase (ROCK) inhibitor; Has been used to increase the proliferation and reduce apoptosis of progenitor cells grown in vitro.
A 83-01
A 83-01 is an inhibitor of TGF-β type I receptor ALK5, the Activin/Nodal receptor ALK4, and the nodal receptor ALK7.
References:
[1] Aliya Fatehullah, Si Hui Tan, Nick Barker, et al. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016 Mar;18(3):246-54.
[2] Marina Simian, Mina J Bissell. Organoids: A historical perspective of thinking in three dimensions. J Cell Biol. 2017 Jan 2;216(1):31-40.
[3] HansClevers. Modeling Development and Disease with Organoids. Cell. 2016 Jun 16;165(7):1586-1597.
[4] Madeline A Lancaster, Juergen A. Knoblich. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014 Jul 18;345(6194):1247125.
[5] Claudia Corrò, Vivian S.W. Li, et al. A brief history of organoids. Am J Physiol Cell Physiol. 2020 Jul 1;319(1):C151-C165.
[6] G R Martin. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7634-8.
[7] Sato T, Vries RG, et al. (2009). “Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche.” Nature 459(7244): 262–265.
[8] Elisa Suarez Martinez , Amancio Carnero, et al. 3D and organoid culture in research: physiology, hereditary genetic diseases and cancer. Cell Biosci. 2022; 12: 39.
[9] Chengyong He, Shaohua Ma, Zhenghong Zuo, et al. Black Phosphorus Quantum Dots Cause Nephrotoxicity in Organoids, Mice, and Human Cells. Small. 2020 Jun;16(22):e2001371.
[10] Mo Li, Juan C Izpisua Belmonte. Organoids-Preclinical Models of Human Disease. N Engl J Med. 2019 Feb 7;380(6):569-579.
[11] Mariangela Scalise, Fabiola Marino, Daniele Torella, et al. From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish. Int J Mol Sci. 2021 Dec; 22(24): 13180.
[12] Xialin Nie, Zhixing Liang, Linsen Ye, Yang Yang, et al. Novel organoid model in drug screening: Past, present, and future. Liver Research 5 (2021) 72-78.
[13] Chen Liu , Chaoyang Sun , et al. Drug screening model meets cancer organoid technology. Transl Oncol. 2020 Nov; 13(11): 100840.
[14] Hanxiao Xu, Kongming Wu, et al. Organoid technology and applications in cancer research. J Hematol Oncol 11, 116 (2018).
[15] Lisi Zeng, Shuzhong Cui, Shengwei Jiang, et al. Raltitrexed as a synergistic hyperthermia chemotherapy drug screened in patient-derived colorectal cancer organoids. Cancer Biol Med. 2021 Mar 12;18(3):750-762.
[16] Maarten H.Geurts, Jeltevan der Vaart, HansClevers, et al. The Organoid Platform: Promises and Challenges as Tools in the Fight against COVID-19. Volume 16, Issue 3, 9 March 2021, Pages 412-418.
[17] Jelte van der Vaart, Mart M. Lamers, Hans Clevers, et al. Advancing lung organoids for COVID-19 research. Dis Model Mech. 2021 Jun 1; 14(6): dmm049060.
[18] Soumya K Kar, et al. Organoids: a promising new in vitro platform in livestock and veterinary research. Vet Res. 2021 Mar 10;52(1):43.
[19] Yaqi Li, Guoqiang Hua, et al. Organoid based personalized medicine: from bench to bedside. Cell Regen. 2020 Dec; 9: 21.
0 notes
Text
Earlier this year a girl from my church asked me to help her prepare for an abortion debate her high school class was going to have the next day, so I quickly threw together some common pro-abortion points and the science and reasoning behind the pro-life position. Since then, a few other people have found it helpful, so I thought I would share it here as well.
_ _ _
1. Biologically, is a fetus a human?
Yes, it comes from two human parents and has human DNA. It is not a parasite because parasites are a different species than the host. It is not just a clump of cells or just a part of the parents like the mother's organs, the egg cell, or the sperm cell - because a fetus has unique DNA different from the mother's and father's DNA, making it an individual human organism.
"Although life is a continuous process, fertilization is a critical landmark because, under ordinary circumstances, a new, genetically distinct human organism is thereby formed.... The combination of 23 chromosomes present in each pronucleus results in 46 chromosomes in the zygote. Thus the diploid number is restored and the embryonic genome is formed. The embryo now exists as a genetic unity."
[O'Rahilly, Ronan and Muller, Fabiola. Human Embryology & Teratology. 2nd edition. New York: Wiley-Liss, 1996, pp. 8, 29.
(More quotes from scientists on the human life cycle and it's beginning at fertilization: https://www.princeton.edu/~prolife/articles/embryoquotes2.html )
2. Does it really matter if it is human or not? "It is so small / it doesn't look human yet / it won't feel pain / it won't know it is alive / etc."
Yes. Again, from a biological standpoint, a fetus is just one of the many stages of the human life. We all begin as zygotes (fertilized eggs), and rapidly begin growing and developing into embryos, then fetuses, then at 9 months we reach birth and begin the infant stage, continuing to become toddlers, adolescents, and finally adults. Just like it is wrong to say "a toddler is less of a human than an adult," it is also incorrect to say "an embryo is less of a human than a newborn infant." Just because someone is less developed than someone else does not make them less of a person.
By 6 weeks (1.5 months/ 42 days since fertilization) the human nervous system is established, and by 8 weeks (2 months) we have observed fetuses to physically react to and recoil from pain. https://lozierinstitute.org/fact-sheet-science-of-fetal-pain/
Sentience (or self-awareness), the most basic ability of something alive to understand its own existence - often first measured in animals by scientific researchers as the ability to recognize themselves in front of a mirror - doesn't develop in humans until months after birth. Everyone knows it would be silly to argue that it's okay to terminate the life of a newborn because it isn't self-aware yet, so logically it shouldn't be acceptable to argue that in the case of abortion either.
3. But abortion is legal / it's none of your business what other people choose!
We can see that many atrocities were legal at one point or another in history. Slavery used to be legal, child marriages used to be legal, many genocides (including the Holocaust) were legal in their countries. Just because something is legal does not make it morally justifiable.
It is always other people's business when injustice is happening. Is it wrong for a group of humans who have done nothing wrong to be intentionally killed, just because they are unwanted? Does that describe abortion? Then abortion is our business.
Bonus: A very technical scientific timeline of human development before birth: https://embryology.med.unsw.edu.au/embryology/index.php/Timeline_human_development . Pro-life is pro-science.
_ _ _
Because of the time restriction, as you can see I did not address the other common pro-abortion arguments centered on bodily autonomy/rape/health of the mother. Those can each be meticulously analyzed and flushed out, and have been many times, but for today I would just like my pro-choice friends to sincerely consider the question of "does your scenario justify ending an innocent person's life?" If you find yourself answering yes, I applaud your logical consistency, but I find how little you value human life rather reprehensible. However if you hesitate, or find offense with the question, consider the evidence and examine your values. You don't need to give an answer now, or take a stance immediately. Give yourself space to study the issue with intellectual honesty and integrity. Don't allow yourself to defend something that you do not fully believe is the right thing.
🖤
73 notes
·
View notes