#what is meant by bioinformatics?
Explore tagged Tumblr posts
Text
DBT JRF 2025: Everything You Need to Know About the Upcoming Exam on May 13
Aspiring to pursue a career in biotechnology research? Then the DBT JRF 2025 is the gateway you've been waiting for! Conducted by the Department of Biotechnology, Government of India, this prestigious exam opens the door to Junior Research Fellowship in biotechnology and life sciences across top research institutes. In this blog, we’ll take a deep dive into everything from the DBT JRF 2025 exam date, eligibility, syllabus, fellowship amount, previous year question papers, and more. Whether you're a first-timer or a repeater, this complete guide is designed to help you stay ahead of the curve.
What is DBT JRF?
Let’s begin with the basics. The DBT JRF full form is Department of Biotechnology Junior Research Fellowship. It is a national-level fellowship program conducted by the Biotechnology Eligibility Test (BET) under the Biotech Consortium India Limited (BCIL) on behalf of the Department of Biotechnology.This exam is meant for postgraduate students in Biotechnology and allied subjects who want to pursue a Ph.D. or research in premier Indian institutions.
DBT JRF 2025 Exam Date
Mark your calendars! The DBT JRF Exam Date 2025 is scheduled to be held on May 13, 2025. Candidates must stay updated through the official website to avoid missing important deadlines related to the DBT JRF 2025 application form, admit card, and result announcements.
DBT JRF 2025 Application Form
The DBT JRF application form 2025 is expected to be released in February 2025. Eligible candidates should complete the form online before the last date. You’ll need to upload your educational certificates, photograph, and signature, and pay the application fee to confirm your submission.
Keep these things ready before applying:
Scanned photograph and signature
Postgraduate degree certificate
Category certificate (if applicable)
Valid email ID and phone number
Don't wait till the last minute – filling the DBT JRF 2025 application form early ensures fewer technical issues and better preparation time.
DBT JRF Eligibility 2025
To apply for DBT JRF 2025, you must meet the following eligibility criteria:
You must hold an M.Sc./M.Tech/M.V.Sc. degree in Biotechnology or related fields.
Final-year students can also apply, provided they submit proof of qualification at the time of the interview.
There is no age limit for appearing in the exam, though some institutions may have their own age criteria for admission.
Ensure you double-check the official DBT JRF eligibility 2025 notification to confirm the subject list and minimum marks required.
DBT JRF Syllabus 2025
The DBT JRF syllabus 2025 covers core areas of Biotechnology and Life Sciences. The exam consists of two parts:
Part A: General science, math, reasoning, and analytical ability
Part B: Specific questions based on Biotechnology and allied subjects
Topics include Molecular Biology, Immunology, Cell Biology, Genetics, Microbiology, Biochemistry, Bioinformatics, and more.
Pro tip: Go through standard reference books and coaching material, and make a detailed subject-wise plan to cover the entire DBT JRF syllabus 2025 in a systematic way.
DBT JRF Admit Card 2025
The DBT JRF admit card 2025 will be available for download around the first week of May 2025. You’ll need your registration ID and password to access it. Make sure you check all the details like your exam center, timings, and photo.
Take a printout and carry a valid ID card along with the admit card on the exam day.
DBT JRF Exam Pattern
The DBT JRF exam is conducted online and comprises 100 questions divided into two sections. You’ll have 3 hours to complete the paper. There’s negative marking in Part A, so accuracy matters!
Part A: 50 questions (General aptitude, analytical ability) – 1 mark each
Part B: 50 questions (Biotech subjects) – 3 marks each
Total Marks: 150
Time Management + Smart Guessing + Revision = Success
DBT JRF Previous Year Question Papers (PYQ)
One of the best ways to boost your preparation is by solving DBT JRF previous year question papers. These papers help you understand the exam pattern, difficulty level, and key focus areas. Try solving at least 5 years’ worth of DBT JRF PYQs under timed conditions to get a real feel of the exam.
You can find the DBT JRF previous year question papers online or access them via trusted coaching institutes like IFAS that provide detailed solutions and analysis.
DBT JRF Fellowship Amount
Cracking the DBT JRF 2025 doesn’t just earn you a title—it brings in financial support for your research journey. Here’s what you’ll receive:
Fellowship Amount: ₹31,000 per month (for the first two years)
HRA: As per institutional norms
Senior Research Fellowship (SRF): ₹35,000 per month after two years, based on performance review
This generous DBT JRF fellowship amount allows scholars to focus on their research without financial stress.
DBT JRF Result 2025
The DBT JRF 2025 result date is expected to be around June or July 2025. Results will be announced online, and candidates can check their DBT JRF result 2025 by logging in with their credentials. Those who qualify will be awarded Category I or II fellowships, based on merit.
Category I: Eligible for Ph.D. admission with fellowship
Category II: Eligible for Ph.D. admission without fellowship, can seek other funding
How to Prepare for DBT JRF 2025?
With the DBT JRF 2025 exam date approaching fast, here’s how you can create a smart and effective strategy:
Understand the syllabus and weightage of topics
Create a timetable covering every subject and revision
Practice DBT JRF PYQs and take mock tests
Revise weekly and focus on concept clarity
Join an online coaching program if you need guidance
IFAS offers structured courses, regular test series, and study materials designed specifically for DBT JRF aspirants.
The DBT JRF 2025 is your chance to step into the world of scientific research with full funding and academic support. With the exam date set for May 13, 2025, now is the perfect time to plan your strategy, revise the DBT JRF syllabus 2025, fill the DBT JRF application form 2025, and aim for success.
Stay connected with trusted platforms and official announcements so you don’t miss the DBT JRF 2025 result date, admit card updates, and more. And remember, smart work always beats hard work when it’s backed by consistency and the right guidance.
0 notes
Text
Who is Elena Lusi?
When reading fringe science that you're uncertain of, one good thing to check sometimes is who the researchers are. Is the off-the-wall evolutionary-genetics paper you're reading from a well respected evolutionary geneticist with decades of experience who's uncovered something deep and amazing? Or is the author a physicist who's Dunning-Kruger ignorance of evolutionary genetics leading them down the garden path while their scientific background makes them sound convincing?
Well, to quote her ResearchGate page:
Elena Angela Lusi is an M.D., PhD. Clinical Immunologist. In 2015, she was the first author to describe that food high in Nickel were linked to obesity in women. She also discovered previously unknown infectious agents that are transmissible and start cancer. These agents are potent carcinogenic biological entities that affect humans and animals in the wild. Immunological strategies aiming at their neutralization will be the key drivers of cancer eradication.
She's a medical doctor (clinical immunologist), as well as having a PhD, who works for a private hospital group. I'm not sure what her PhD was on, as I can't find any records of it, but most of her early work appears to be regarding T-cells and HIV. So far so...well it's a little bit of a branching out for someone working as clinical immunologist but y'know, it's not like she has a completely inappropriate background. Kinda.
But if you go through her research history, something happens around 2018. She's been studying the effect of nickel on weight in women but she's started publishing about finding giant viruses in humans in the journal F1000 Research. A paper titled Discovery and description of the first human Retro-Giant virus, which much like the CTVT work, big if true.
F1000 Research is an interesting journal in that it's not the standard submission > peer-review > publication format. Instead, authors submit a manuscript, which is checked over by the editorial team and then published online, where it can be commented on and openly peer reviewed, and then revised by the authors, with prior versions retained and publicly visible.
So far, one person has positively peer reviewed the aforementioned, the now disgraced scientists Didier Raoult. But what's more interesting to me is the comments section, which, uh...whelp they don't paint a great picture of Lusi's scientific approach. It's linked and I encourage you to go read it but I'm going to copy-paste the exchange under a cut here for those who don't want to go link hopping.
Suffice to say it starts off with a very reasonable question from someone with knowledge of the field and research methods involved. Her response, however, starts of seeming reasonable before descending into all caps and putting the very concept of bioinformatics on blast.
My favourite, extremely scientific part was
BOTTOM LINE > THE HUMAN GIANT INFECT AND CAUSE CANCER> FULL STOP.
Very science, much logical argument.
To be clear, this isn't meant to be a character assassination of her. I'm very much trying to assess the science but the actions of a person around their research can still be valuable in situations like this. Being an arsehole doesn't make your data invalid. But it may help eludicate how objective someone's work is vs. glory hunting or irrational emotional attachment to a pet theory.
Reader Comment 29 Jul 2019 Daniel Elleder, Czech Academy of Sciences, Czech Republic The interpretations of the sequences obtained in this work are quite confusing and I think incomplete. E.g., the main 150-bp amplicon does not seem to be related to retrovirus sequences. The tree in Fig4 also does not prove any relatedness, the position of the 150-bp sequence is an outer branch without any support. However, this sequence has very close hit in human genome (chromosome 4), which I do not see mentioned. The long contig (first position in supplementary tabe, contig 1) has basically identity to E Coli chromosome. The hit described as gag-akt fusion (contig 1432), and probably used to argue about relatedness to retroviruses, has similarity to gag-akt fusion in GenBank, but in the akt protein region. So it is probably of human origin, no retrovirus sequence involved. D. Elleder
Author Response 05 Aug 2019 Elena Angela Lusi, St Vincent Health Care Group, University College Dublin, Dublin, Ireland
Dear Daniel Elleder Thank you for your comment. A version 3 of my manuscript, with updates, will be released shortly. In this new version, I will present further experiments including the isolation of the giant particles from human cells, their purification on a sucrose gradient, whole genome sequence (this time repeated in duplicate), additional EM analyses, transformation assay on NIH 3T3 cells after giant particles infection and tumours formation in mice.
The giants induced peritoneal metastatic disease after three weeks post infection. Please, See links below. Unfortunately, especially in the light of the new data and metastatic disease in mice, your comment is neither here nor there.
In this letter, I will anticipate some of my results to address fundamental concepts that seem to be missing.
1. The giants are not typical viruses, but microbial cell-like entities. They have genes in common with the three domains of life: eukarya, bacteria and archaea. Like bacteria, they retain the Gram stain.
2. The reported particles have been extracted and purified through a sucrose gradient, https://drive.google.com/open?id=1ai8MpZ0eat5a7LT7b43NPXNRWy0McJwo
3. These human giants infect and transform NIH-3T3 cells in vitro, https://drive.google.com/open?id=1a6ofZQNpxaZElAtCbX5SVpPdQxGAo1h2
4. The giants induce tumours formation in mice, https://drive.google.com/open?id=1Xbxtf1ximI3oTJsA3I8-1dq8CdO8I8XX
5. The tumours formation in mice is REAL and NOT a bioinformatics score.
6. Why do my Giants cause cancer? Simple : they have oncogenes INSIDE their mega-genome!
7. The giants fulfil the Koch’s postulates. I have re-extracted the giant particles from cells that became cancerous.
8.The giant have RT activity. Again this is a REALITY, a real biochemical reaction and not ONLY a bioinformatics annotation.
9.Bioinformatics are prediction and they CANNOT replace the biological reality. Metagenomics does not replace the need to cultivate and isolate microbes.
10. The function of bioinformatics is predicting, not explaining. Koch or Pasteur discovered their microbes without any software. Rous discovered RSV in 1911. ....And the databases grew, and everyone annotated their data by searching the databases, then submitted in turn. No one seems to have pointed out that this makes your database a reflection of your database, not a reflection of reality.
At this stage, with the striking evidence of tumours formation in mice, your debate on the 150 bp fragment is trivial . Honestly, I don’t base the discovery of this magnitude on a single 150 bp fragment. This was just a preliminary RT–PCR following an old protocol commonly used in retrovirology. Please refer to the new data and whole genome sequence. If the tagmentase enzyme disturb you with some background noise, just take the E. Coli sequence off . The results doesn’t change: a mega-genome is confirmed (https://mega.nz/#F!7dID0aSa!8bA-4qVdPeiY0tsSbd8G7g).
The predominant archaea features coexist with the presence of an anchoring system typical of viruses and gag-pol proteins of oncogenic retroviruses complete their chimeric essence. The most valid bioinformatics tool was achieved through Blast2GO. This simulation went really close to the real life scenario (example of realist and desirable bioinformatics). The annotations of the many oncogenes in the mega-genome proved to be true, since the giants induced cancer in mice (please see the cancer in my mice). In addition, the predicted transforming gag-pol genes matched with the retroviral antigenicity, documented at EM immunogold and RTactivity .
Note that , in my experiments, a filter supernatant DOES NOT transform.
These human giants was missing because of our definition of viruses and some people seem to struggle (new concepts , new paradigms, dynamics) .
After inspecting my data in the provided links, I would like you to answer, just with a YES/NO, my questions:
What are the purified structures depicted at EM? Are they something human? If yes, explain why .
Do these Giants INFECT and transform NIH 3T3 cells?
Do they induce peritoneal metastasis in mice?
Do they reverse transcribe in a real biochemical reaction?
Do they retain the Gram stain?
Do they have a mega-genome?
Can they be isolated every single time just with a routine sucrose gradient by independent operators?
At each extraction , do the EM reveal the same dimension and structures?
Do they have multiple cell based oncogenes? (BISHOP)
Do they have a capside?
Does a filtered supernatant do the same?
Is the anti-Felv gag antigenicity of the particles in line to the PREDICTED akt-gag and the documented RT activity?
Does bioinformatics replace biology?
Are you familiar with the concept of TRUC and cell like entity with genes in common to the three domain of life?
Is it true that viruses carry also fragments of human sequences?
Is it true that the transforming gene product, P70gag-actin-fgr, of Gardner-Rasheed feline sarcoma virus (GR-FeSV) is a single polypeptide composed of regions derived from cellular and viral genes?
What is MPGNL?
Did you see cancer in my mice?
Would you like to be treated for a physiological illness by a physician who is not sure that there are human bodies, and who uses information systems that lack real referents?
BOTTOM LINE > THE HUMAN GIANT INFECT AND CAUSE CANCER> FULL STOP.
The retroviral nature is peculiar, but it is just a detail. The giants are much more: an entire cancer factory that transform in few days. Try with your bioinformatics skills to discover a new sequence. Not a variant or a subtype of something already known, but something like a TRUC that induces cancer in your chicken in three weeks. Without having your skills, I was able to achieve this in mice. My giants might open the door to a preventive vaccine against cancer, since they give possibility to target an entire shuttling system of oncogenes and not just a solitary molecule involved in carcinogenesis. It is like discovering HPV for the first time, or EBV for the first time, just to mention few.
This time is not a virus, but a TRUC : Not an oncogenic virus, or a slow virus, with few proteins that you can count on the fingers of one hand, but an infectious oncogenic CELL-LIKE MICROBIAL ENTITY that looks like a bacteria, with hundreds of proteins, carrying in its large mega-genome a transforming arsenal. A sort of small autonomous infectious cell, a simplified version of its eukaryotic counterparts, specialized in carcinogenesis.
The discovery of this microbial entity with an acute transformation mechanism and infecting humans suggests that the number of cancer of infectious origin would be even greater than what is supposed.
In my discovery there are no shadows, no quarrels, no misconducts and no thefts. I feel blessed and the Retro-giants is a gift.
Suggested references
"Anti-realist bioinformaticians work with data handed over to them by realist biologists. Some features of some information systems can be improved if bioinformaticians became realists as well. Still another thing makes it desirable that bioinformaticians become realists: they could then possibly provide feedback to biologists". https://www.sciencedirect.com/science/article/pii/S153204640500078X Elena
CTVT and a weird niche theory I fell down the rabbit hole about - giant transforming retroviruses???
This is a story about how a single line on a wikipedia page sent me down a rabbit hole of finding one scientist's fringe theory that's juuuust plausible enough to make me question everything while almost certainly being absolute fucking bunk.
Some background
So, on parts of tumblr at least we all know about Canine Transmissible Venereal Tumour, aka The Immortal Cancer Dog. For those who don't know, it's a cancer dogs get, usually on their junk, that unlike most other cancers, isn't made up of their own cells. The cells are actually all descended from this one dog or wolf that lived like 11,000 years ago and are, arguably, all technically that one dog. A dog that became a single-celled infectious disease.
We have a wealth of genetic, histological and observational evidence for this. As in, we know it what population of canids it came from, we know it's got a weird chromosomal structure compared to normal dogs, we know it's genetically distinct from the hosts. We also know it's not the only one out there: There's a similar thing in Syrian hamsters and also the famous Tasmanian Devil Facial Tumour Disease (DFTD).
Which made me pause when I was reading something on wikipedia about the devil facial tumour and saw a line mentioning that it was now known to be caused by a giant virus, much like CTVT. Which...huh? Oh I hadn't heard that afore.
Giant viruses
Ok so giant viruses are a thing and they're fuckin cool. They're a relatively recent discovery and comparatively huge, i.e. bigger than a bunch of bacteria. They were only discovered in 1981 and we still don't know an enormous amount about them but they're big and have large genomes and because of the way viruses are they're not easy to detect unless you're specifically looking for them.
They show up under microscopy (sometimes) and you can find them with genetic probes but you gotta already be looking for them to see that really. Current research though basically says they're more common than we think, just overlooked, and there's software out there that scans through genomic data to find sequences that might indicate their presence. There's even a possibility that one group might be involved in some cases of pneumonia in humans, though I need to stress that that's extremely not confirmed right now.
The "wait, what?" moment
So I mentioned that it was a line in the wiki article for DFTD that had me going "wait, really?", the line in question was this:
A study found evidence for an infectious agent resembling a giant virus that was capable of turning heathy cells into cancer cells. It was found to be a huge retrovirus with similar viruses being found in human and canine cancer cells.
Big If True.
So of course I check the source, which was a 2020 paper by Lusi et al. titled "A transforming giant virus discovered in Canine Transmissible Venereal Tumour: Stray dogs and Tasmanian devils opening the door to a preventive cancer vaccine".
Hang on, CTVT not DFTD? This is where some alarm bells went off because uh, as mentioned at the start, we know a shit ton about CTVT. Including the fact that it's all one specific dog. Which doesn't fit at all with the idea that it's caused by a virus transforming host cells into cancer cells.
So what fucking gives? What is this research that fully overturns decades of pretty conclusive research to the contrary?
Is this another case of Dr Barbara McClintock? Who spent decades being ridiculed by the scientific community over her wild theory that was, in fact, 100% right even if it seemed to fly in the fact of all prior evidence?
Or is this a Dr Donald I. Williamson situation wherein a scientist with appropriate training is just wildly but extremely vehemently wrong?
#science#genetics#cancer#canine transmissible venereal tumour#devil facial tumour disease#niche science#stay tuned for more
34 notes
·
View notes
Note
i really want to go into botany and/or mycology but i’m physically disabled and wouldn’t be able to go on hikes like you do. do you think i should try to find something else to study that doesn’t require me to go to wheelchair inaccessible places?
This is a great question. I used to do regular field seasons until I was diagnosed with a chronic illness that limits how much fieldwork I can do, so this is a topic I’ve thought a lot about as I’ve had to shift the way I do things.
First, I encourage you to think about what you’d like to do on a day-to-day basis. Do you want to be interacting with the organisms you’re researching, or would you be happy working with data at a computer? For me, this has meant focusing more on learning bioinformatics, but for others it could mean getting into stats or modeling (this is especially useful if you’re more interested in ecology). If you’re more interested in working with the organisms, you have options of pursuing avenues of research that are more lab- or greenhouse-based (more often than not those places aren’t super accessible but accommodations can and should be made).
Next, remember that you deserve to research whatever you damn well want regardless of your physical ability, and sometimes it’s necessary (unfortunately) to self-advocate and demand that you’re accommodated. When I came into my graduate program, I made it clear that I couldn’t do a lot of fieldwork, and I’ve been lucky to have motivated undergrads collecting most of my specimens for me. It’s absolutely exhausting to have to self-advocate all the time, or to deal with microaggressions on a regular basis, and it’s up to you to decide if it’s worth it.
Lastly, I personally get a lot out of engaging with community and knowing I’m not alone. I’ve recently discovered a strong community of disabled academics on Twitter, and seeing other people with similar issues who know their worth and are actively working toward their goals can be inspiring. On the flipside, having people to talk to when you need advice/support is invaluable.
Let me know if you have any other questions! This isn’t something I’m super well-versed in but I do have a lot of thoughts on the matter anyway.
668 notes
·
View notes
Text
Release Me Into Orbit
(Dark!Bucky x Black!Female Reader)
Summary: Bucky and the Reader are trying to heal from the trauma of their pasts.
A/N: Here we dive into the past. The true beginning to our characters stories. This story takes place both in the past and the present so get ready for that. I do plan on releasing the next Chapter of Invisible Chain soon! Stay tuned.
Warnings: Non-Con, Dub-Con, Violence, major character death, Manipulation, emotional abuse, physical abuse, eventual Kidnapping, Breeding Kink, and angst etc later in the story.
Honestly More tags will be added.

Ch 2: 2014
Longing.
It was hard for Bucky to see anything through the small opening in his cell door. He had lost count of how many years he had been locked up a long time ago. There were no windows that allowed him to even guess when it was night and when it was day. Inside his cage they made sure he had no human interaction; they left Bucky completely alone. As the years passed the only thing keeping him company was his own thoughts. Bucky would try his hardest to focus on his fragmented memories before the war. Those memories contained the few good things he could remember about being human. His first kiss behind Sal’s Diner. The smell of his mother’s home-made cherry pie. The scary thing was he couldn’t even remember his mother’s face, just the smell of her fragrant pie. To be honest he couldn’t remember any faces. They were all just blurs. When it came to his captors all he knew is that they had the same routine in place for him every day. They would feed him his one meal, then they would return to remove him from the cell. The strong ones would drag him through a maze of corridors until he reached a room filled with bright fluorescent lights. The lights illuminated the dreaded device they used for their experiments.
Rusted.
The experiments were the only time he interacted with anyone outside of his cell. Bucky could never see them properly as they always hid behind the bright lights. All he ever got to see were their shadows as they moved about speaking their foreign tongue. Nobody ever spoke to him unless they were reciting the very words that caused him great mental distress. Those words were a curse that was cast on him a long time ago after the war. One that followed him, captor to captor. They would often drug him, just enough so he’d be a little disorientated, but they made sure he’d still be able to feel and hear everything. Once drugged, that’s when they would begin torturing him. The only thing they would change in his routine is the method in which they tortured him. Each time they would try a new method to test the limits of his body and the limits of his mind. Unfortunately for Bucky, the day didn’t end until he physically and mentally could not take anymore. Eventually, they didn’t even need to use pain or the dreaded spell to get him to comply.
Seventeen.
It was a strange sensation to not be in control of one’s own body. It was a sensation that Bucky should have grown used to, but he never could. It was as if his own consciousness was taking a back seat while someone else controlled him over and over, each driver just as brutal as the next. He was always vaguely aware of what atrocities they made him commit in the name of science, and that in itself was torture to live with. When they would return him to his cell, he could barely get a wink of sleep as the images would replay over and over in his mind. Their screams ringing in his ears. This was what they had reduced him to, he was just a tool to them. Eventually, he had to do everything in his power to keep himself from going completely mad and that just meant numbing himself to it all. The bright-eyed man from Brooklyn that was just trying to save the world was gone years ago. In his place was a murderer. A weapon. A monster. And that’s all he’d ever be.
Daybreak.
It was a day like any other, except it was storming outside. Bucky could tell by the sounds of the fierce winds howling against the facility that and he had finally been moved to a cell with a small window. Bucky had learned that he had been with these particular captors for several years. Honestly, everyone that ever ‘owned’ him was the same in his eyes, so it didn’t matter how long he was kept. Eventually, he’d just be handed off or kidnapped again by someone else that wanted him. Luckily for him, years of compliance and loyalty earned him an upgraded cell and some limited freedoms. Along with some of the smaller changes he was finally able to speak their tongue, just another language to add to his impressive portfolio. There weren't a lot of things Bucky could do to entertain himself. If he wasn’t on an active mission, he was pretty much tied to his room unless they allowed him a break from his cell. In his free time, all Bucky could do is entertain himself by working out. And that’s all he ever did.
Furnace.
He was face down as his palms supported his weight in a push-up position.
“Two hundred and Forty-Nine.”
“Two Hundred and Fif-”
Nine.
He suddenly paused looking towards the door of his cell. He could have sworn he had heard a voice. Bucky waited patiently, but there was only silence in return. Perhaps he had imagined it, or maybe he was truly going mad. He returned his gaze to the floor before he heard it again. It was a voice, it was faint, but it was definitely a voice.
Benign.
“Hello?” it called out softly as if in a whisper.
Homecoming.
The voice was suddenly followed by the sound of the metal slot on the door sliding to reveal two eyes peering at him. Bucky was unsure of how to respond, mainly because he was unsure what was going on. The men who came to retrieve him from his cell never spoke to him, and this person was clearly speaking English. English felt foreign to him now, he hadn’t heard it in so long. Bucky suddenly sat up watching the eyes on the other side of the door. Again, there was a moment of silence before they spoke again.
One.
“Can you understand me?”
It was a woman.
Freight car.
---
“Mom! Hurry, I’m going to miss my flight!”
You rushed down the stairs with your bags in your hands. Your shoes caught on the last step almost causing you to trip. A quiet curse left your lips as you attempted to stabilize yourself. Your father watched as he tried not to laugh at your misfortune. Carefully, you handed him your bags and he tossed them into the trunk of his truck.
“Fragile!” you shouted at him.
You crawled into the back of the car as you patiently waited for your parents to join you. Eventually, after what seemed like another 30 minutes gone by, they entered the car. “Do you have everything?” your father asked one last time.
“Yes, now let’s get going!”
It felt like freshman year of college all over again. Instead, you were a graduating senior and you had just accepted an opportunity of a lifetime. A few months ago, your professor, Mr. Brigmova, had presented your class with an opportunity. The top five students in the program would be able to join him in a work-study program. You were still unsure how you beat out several other students, but you did. You felt extremely lucky to be able to partake in such a program. To be among the top 5 students in your area of study was everything you could have hoped for. It showed your parents that it wasn’t a waste of time or money to send you out of state for school.
Ever since middle school, you had always wanted to study genetics and biology. When you reached high school, you learned about Bioinformatics and molecular genetics and you made up your mind on what you wanted to do. When you told your parents, they began immediately running numbers in their heads. Feeling guilty about your career choice and the school you chose to attend; you worked your ass off to get and keep a 4.0 GPA. In turn that promptly led you to the 3rd spot among the top 5 students. And not to toot your own horn, you were the only double major among them. Double the hard work, so deep down you felt as if you were #1 anyway.
As a result of all your hard work, you were flying across the world for the first time. You’ve never been so nervous before your professor had revealed the location, you had never heard of Sokovia. You wouldn’t have been able to locate it on a map either, it was such a small country. This was also the first time you had to travel without your parents, and you’d be away for them for five months. You would have no safety net out there. Before you knew it, you had arrived at the airport and you could feel the butterflies settling at the bottom of your stomach like a heavy rock. You felt like you had to use the bathroom, but you knew better than that. Your parents helped you remove your bags from the car as the other cars honked impatiently. You flipped the respective vehicle off before embracing your parents one last time. You exchanged your goodbyes before you disappeared behind the airport doors.
It was the longest flight you had ever been on. After getting off the plane, you were tempted to kiss the frozen ground, but you had seen enough cartoons to know what would happen. In Sokovia, the winters were harsh around this time, and thanks to your research beforehand you were prepared. Or at least you thought you were. You exited the airport to be greeted by the harsh Sokovian winter.
The bitter cold of Sokovia nipped at your bare cheeks and the tip of your nose. Slowly it seeped into the pockets where you kept your gloved hands numbing your fingers slowly. The cold was nothing like you were used to back in the south. Immediately, regret settled right beside the nervousness as you realized you wished you had stayed behind the airport doors just a moment longer as you waited for your ride.
The streets were not as busy as google images made it seem. It was probably due to the fact that it was unbearably cold, and it was late. You desperately wanted to return to the warmth of indoors, but you feared missing your shuttle. You moved about in hopes that it would warm you up while you waited for the shuttle as you watched it grow dark. Not wanting to expose your hands to the cold you peeked at your pocket for the time before you left you made sure to adjust it for the time difference. The shuttle was running late and that worried you, you knew nothing of the language to get help if you ended up stranded.
Eventually, you took a seat on your suitcase, holding onto it tightly. The last thing you would want is to be robbed in a completely different country. You waited for what seemed like almost an hour in the cold before you saw the shuttle pull up in front of you. Slowly you stood on your two feet stretching as you watched the shuttle door slide open, only to reveal your professor. Mr. Brigmova was a tall man with an average build in his early forties. He had dirty blonde hair and striking gray eyes. If not for the slight wrinkles near his eyes, he could have easily been mistaken for his early thirties. He motioned you inside as he jumped out to grab your bags for you. You did as you were told climbing into the vehicle as the warmth inside embraced you. You watched as Mr. Brigmova carefully placed your bags in the back seat of the shuttle. He flashed you a smile as he slid into the seat next to you. “It is good to see you, Y/N.” He greeted you. “It’s nice to see you too, Mr. Brigmova,” you replied.
“Please, call me Peter. We’re colleagues now.” He responded. You noted it was just you, Your professor, and the Driver. Out of everyone you were the last from the program to make it in. Your professor turned towards the driver tapping the back of his seat, “My gotovy k rabote.” He mentioned in his native tongue. You weren’t completely sure what he had told the driver but the fact that the shuttle started moving probably was related. Peter turned to you noting how tense you were placing a hand on your knee and squeezing it lightly. His action surprised you, but he only flashed you a smile in return,
“Relax you’re in Sokovia now.”
#bucky x you#bucky x reader#dark!bucky barnes#bucky fanfic#dark!fanfiction#Dark Fic#dark!marvel#dark!bucky x reader#dark!bucky x black!reader#dark!bucky x you#dark!bucky x y/n#dark!mcu#black reader#bucky × black reader#sam wilson
57 notes
·
View notes
Text
Prompt Nr. 1 for @onemuseleft: You had an assigned seat next to them at a wedding for a mutual friend. I altered it a tiny bit. Also one day I will write a Guardian fic, but today is not that day (sorry!).
---
Truth to be told, Wei Ying didn’t exactly know what he was doing here. He would have been invested in the proceedings if this was his sister’s wedding, or his brother’s (if it was ever going to happen). But this was the wedding of the daughter of one of Jiang Fengmian’s business partners, and Wei Ying had no idea why her family had thought it necessary to invite so many people. Including Wei Ying, who had absolutely nothing to do with his stepfather’s company.
To make matters worse, his entire family had been divided when they were seated. Jiang Fengmian and Yu Ziyuan sat with other important business partners, while Jiang Yanli was sitting with her peacock husband (UGH). At a third table, Jiang Cheng was seated together with what Wei Wuxian assumed were eligible young daughters of the aforementioned business partners. How either A-Cheng or the daughters escaped, Wei Ying couldn’t tell, but he was internally preparing for a whole lot of complaints once this was over.
And then, there was Wei Ying. He had been seated at a table of mostly men around his age, which he was grateful for. No eligible young misses for him. What he was less grateful for was the fact that most of his seatmates barely opened their lips, and obediently listened to all the speeches and the entertainment without even cringing at the terrible allusions to the newly wed couple’s sex life. Wei Ying had to tune out after a while and studied the faces at his table, instead. There was a group of four to his right that was very average, and mostly stuck out by the fact that they only conversed among each other, giving Wei Ying no opportunity to strike up a conversation. On his left were three men, all dressed in light blue and accents of white, which Wei Ying thought was a rather odd choice, but Jiang Cheng just had had enough time to whisper to him that these were from the extremely wealthy Lan family, and apparently, the colour scheme was a whole thing.
It wasn’t just the clothing that set them apart from the rest of the crowd, however. All three of them were exceedingly handsome, and two of them looked so alike they might have been twins. There was one major difference between the two maybe-twins, however; while one of them had smiled at Wei Ying in a friendly way and introduced himself as Lan Huan as he sat down; the other one looked grave and severe, and had only just said his name (Lan Zhan) before he sat down and kept his silence. And, with Wei Ying’s luck, it had been Lan Zhan whose seat had been directly next to Wei Ying’s. Which meant that he would be without a conversational partner for the entire duration of this banquet.
Finally, finally, they were spared another round of speeches and were served the appetizers. At least he had something to do now, so Wei Ying enthusiastically went for the food. Out of the corner of his eye, he watched the little group of Lan family members to his left. They all ate with elegance and restraint. Every movement looked carefully studied. Was this a model shoot or what? Wei Ying couldn’t believe it. He hadn’t ever seen anyone eat so prettily.
And they just had to put Wei Ying next to these people, he thought uncharitably, so that it would be perfectly evident that he was a boor.
Once the food was finished, Lan Huan carefully wiped his lips with his napkin (there was nothing to wipe off) and smiled at Wei Ying, leaning slightly forward to get a good look around Lan Zhan’s tall figure.
“I understand,” he said in a friendly, conversational tone to Wei Ying, “that you are the adopted son of Jiang Fengmian?”
“Yes,” Wei Ying replied, a little embarrassed that this was still the way in which he was known in these circles: Wei Ying, the unfortunate orphan who was lucky enough to get adopted by an affluent business magnate. “But I have nothing to do with the company.”
“Ah,” Lan Huan said, making an agreeing sound. “What are you doing then?”
“I’m still at university, working on my PhD in Bioinformatics.”
Lan Huan looked slightly impressed at that, and started asking him questions about his research. Wei Ying was relieved. He could talk about his research endlessly, and if there was anyone who was willing to actually listen (no thanks to A-Cheng), he was even happier.
“Oh, you study at Gusu University?” Lan Huan asked when Wei Ying mentioned his lab. “You should have lunch with Lan Zhan sometimes, then. He’s also at Gusu, though he’s in humanities. He’s also doing a PhD.”
“Wow,” Wei Ying said, turning to Lan Zhan and trying not to get discouraged by the forbidding expression on Lan Zhan’s face. “Humanities, huh? I was too stupid for humanities. Too many possibilities. Give me some numbers, I can handle that much better.”
“I am sure Wei Ying is very smart.”
It was the first time since his taciturn introduction that Lan Zhan had spoken at all. Wei Ying blushed at the compliment. He had been called smart before, but all too often, it felt like empty flattery or even a dismissal of who he was as a person, but this… this felt entirely genuine. To say such a thing after holding his silence for almost an hour, Wei Ying couldn’t help but react.
“Thank you. I’m sure Lan Zhan is smarter than me.”
Lan Huan laughed quietly in the background. He turned towards the other member of the Lan family, apparently the cousin of Lan Huan and Lan Zhan, and started speaking to him.
Wei Ying stared at Lan Zhan, trying to figure out what to say next, but now that Lan Zhan had turned to him, he couldn’t help but notice what beautiful eyes Lan Zhan had; strangely expressive eyes that were complimented by a handsome (though invariably serious) face. To have the attention of such an attractive man all focused on himself made him feel slightly embarrassed.
Then he remembered that he was shameless, and he leaned forward with a smile.
“But Lan Zhan, Lan Zhan, if you’re really at the same university as me, we should definitely have lunch! My lab mates are such bores, I tell you. They always eat lunch in the lab! They never go out! So antisocial! I’m lonely!”
At first, he was almost sure that Lan Zhan was going to retreat, going to reject him out of hand, but then, to Wei Ying’s surprise, Lan Zhan hummed in what must be agreement.
“There is a vegetarian takeaway just next to my office,” Lan Zhan said quietly. “It is very good.”
Wei Ying felt his smile get bigger. “Oh, really? You have to introduce me, and tell me all the best orders!”
“Hn,” Lan Zhan agreed.
“Hehe, it will be fun,” Wei Ying enthused, pulling out his phone. “You should give me your number, so I can ask when you’re free!”
Lan Zhan carefully took his phone and entered his number and even his email. When he gave it back to Wei Ying, their fingers brushed. It was an innocent enough touch, but somehow, it felt like Wei Ying had just stuck his fingers into an electrical outlet.
Nervously wriggling in his seat, he looked down at the information Lan Huan had entered.
“蓝湛, huh,” he said once his brain had gone online again. “A very fitting name.”
“Thank you,” Lan Zhan said. Then he kept staring at Wei Ying silently.
“Oh!” Wei Ying suddenly realised. “You want my name? It’s–”
The wedding banquet was over far, far too soon. Suddenly, the last dishes were cleared, the last entertainments were had, and the night was coming to an end. Everyone was getting ready to leave.
But, Wei Ying thought as he took his leave from the Lan family and returned to his own, waiting for him, it had been time entirely well spent.
He kept clutching his phone in the car on his way home, staring at the last message he had received despite Jiang Cheng’s strange looks.
I’ll see you soon, Wei Ying.
85 notes
·
View notes
Text
August 24th-September 3rd
1. Monday, August 24th: What classes are you taking?
Isolation and Separation Methods of Macromolecules, Human Genetics+labs, Molecular Biology Seminar, Methods of Molecular Biology, Bioinformatics, English- Prep for the UniCERT, Regulation of Gene Expression, Bachelor Thesis Seminar (from Biotechnology)
2. Tuesday, August 25th: What time does your alarm go off?
Tomorrow about 5:30 cause I got doctors appointment.
3. Wednesday. August 26th: Do you check your phone when you first wake up?
Yes, everytime.
4. Thursday, August 27th: What is your favorite season? Why?
I mean every season has something in it. I love summer and autumn mostly.
5. Friday, August 28th: Tea or coffee?
Both. Coffee in the morning, tea in the evening.
6. Monday, August 31st: What is your favorite thing about school?
Meeting friends and labs.
7. Tuesday, Sept. 1st: Favorite school memory! (Not online lol).
When I went to the molecular biology lab for the first time.
8. Wednesday, Sept. 2nd: How do you take your coffee?
I'm not quite sure what is meant by this question. But I take coffee in my Starbucks bottle. (idk if u meant that) :D or if u mean how often I drink it I mean 1 coffee/day
9. Thursday, Sept. 3rd: How do you take your tea?
same as coffee (8.)
@spell-studies
1 note
·
View note
Text
10 best Summer Schools In India
1.Symbiosis Summer School: Meant for students of class X, XI and XII. Duration 6th May to 27th May 2019. Students can choose one module from available options: (Each module will include a mix of sessions from the different SIU UG institutes)
Law + Economics + Liberal Arts Design + Media + Photography Business +Computer Studies + Engineering Health Sciences + Nursing + Biomedical + Culinary Fees: INR 120, 000 (including accommodation and meal) Website: http://www.symbiosissummerschool.in/programm
2. The Young Scholars Programme at Ashoka University: A unique summer residential programme which introduces high school students to the idea of a liberal arts education. This is an excellent program for outstanding high school students in Class 11 and 12
Dates: Two-week programme Location: Ashoka University, Sonipat (Haryana) Application Fee: INR 500/- to be paid along with submission of application form Programme Fee: INR 40000/- (inclusive of taxes) Need-based scholarships available For questions: [email protected] https://highschool.ashoka.edu.in/Login.aspx
3.Jindal Global Summer School: It’s a 2-week program that offers a unique opportunity to have fun, learn new things, make friends and in the process embark on a great voyage of self-discovery. Students who are currently pursuing class 9,10,11 and 12 are eligible to apply regardless of the board or school they are studying in.
Courses on offer are · Business Management, Leadership and Entrepreneurship · Media, Communications & Public Affairs · Law, Justice & Democracy · Diplomacy, International Relations, and Peace · Economy, Public policy, and Development · Liberal Arts, Culture, and Humanities · Architecture, Planning and Sustainability · Banking, Finance and Accounting
Location: Sonipat, Haryana Fees: Website: http://www.jgu.edu.in/event/jindal-global-summer-school-aspire-india-scholarprogramme-high-school-students-ixxxi-xii
4. Amity University Summer School Programme: Amity University offers two-week summer programs in Science & Technology, Management, Communication, Creative Programmes, Law, Hotel Management, Travel & Tourism, Psychology, and Foreign Languages.
Dates: 27th May 2019 to 7th June 2019 Location: Noida Fees- Course Fees: Rs. 10,000/- (Fees subsidized by Amity Youth Found). Hostel Fees: Rs. 4,000/- (Including Breakfast / Lunch / Dinner). Website-http://www.amity.edu/summerschool/programmes.asp
5. Young Leaders for Active Citizenship: Founded by Harvard and Oxford alumni in 2016. YLAC aims to increase the participation of young people in the democratic process and build their capacity to lead change.
The high school achievers program is one of its kind. It is designed to help young adults become active agents of change in their communities. The program will take place at three cities that is Delhi, Mumbai and Bangalore.
Fees: 20,000(depending on the modules).Scholarships are provided based on merit and the amount ranges between 25% to 75% of the total program fee. Website- https://ylacindia.com/
6. Summer Immersion Program at Flame University: If you are a high school student between the ages of 15-18 or in grades X, XI, XII, FLAME University’s Summer Immersion Program is the ideal place for you to invigorate your mind and imagination, to figure out what interests you, and to strengthen your skills through various academic and non-academic modules.
The modules that will be covered in the program include:
Critical Thinking
Reading Literature and Writing
Contemporary Issues and Debates
Public Speaking and Debate
Painting and Sculpture
Introduction to Cultural Studies
Environment Studies with Fieldwork
Film Appreciation
Leadership and Group Dynamics
Sports (Tennis, Football, Basketball, Swimming, Badminton, Cricket)
Organic Farming
Date of the program- Batch 1 – May 12, 2019 – May 25, 2019, Batch 2 – June 30, 2019 – July 13, 2019
Fees- INR 50,000/- which includes tuition, course materials, meals, on-campus accommodation and service tax. Website-https://www.flame.edu.in/academics/summer-immersion-program.
7. Young Technology Scholars: is a two-week intensive summer program where students are exposed to real-world engineering and problem-solving. The program curriculum has been designed by senior faculty from leading universities of the world, in collaboration with alumni from top institutes like Stanford, UC Berkeley, HBS, IITs, and the IIMs. It’s a great program for the kids who want to explore the scientific, creative (design) and business aspects of AI, Robotics, Electronics, Optics, Data Sciences, and Bioinformatics.
Location- Gurgaon. Program Date- 1st June 2019- 14th June 2019. Fees: Program fee for Early Bird applicants: Rs 75,000. The program fee for Final Round applicants: Rs 90,000. Scholarships (up to 100%) are available for students, on need basis. Website-https://youngtechscholars.org/
8. Doon’s Summer School Program on Leadership: An intensive but lively and enjoyable programme based on case studies, creative experiential learning, design thinking, problem solving, outdoor adventure, social service, self-reflection and group interaction, guides students through modules drawing on The Doon School’s immense experience in developing influencers, social activists and thought leaders. The Summer at Doon programme is open to interested students aged 14-18 years in Grades 9-12.
Admission Criteria: Interested candidates are required to provide the following:
A completed application form obtainable from the website: www. doonschool.com/summer.doon/leadership A short personal essay A reference from a teacher Payment once acceptance has been acknowledged The course cost is Rs. 1,45,000/- . This is inclusive of everything apart from pocket money and transport to and from school. Location-Dehradun Website-http://www.doonschool.com/summer-at-doon/
9. Summer Programme at UWC Mahindra College: The summer programs at the UWC Mahindra College is excellent programs to experience social awareness and multicultural diversity. The programs are usually 1 to 3 weeks long. If you want to undergo experiential learning within an international community while focussing on real-world issues (waste management, food & agriculture, sustainable human habits, religion, gender equality, politics etc.), then this is a must-attend the summer program.
Following Programmes are being offered in 2019:
YES: Youth, Environment & Sustainability (1 – 9 June 2019) TGIF! Theatre, Gender, Identity, Film (9 – 22 June 2019) Encounter India (22 June – 14 July 2019)
Location: Various locations in India (mostly rural and semi-urban) Fees: INR 31, 000 to 193, 000 (depending on course modules and duration) Website: http://uwcmahindracollege.org/join-us/become-a-summer-student
10. India Summer School by King’s College London: In addition to the summer programs in London, King’s College London (in partnership with Lady Shri Ram College) also offers summer programs in India. You could study International Relations or Marketing Management with International Marketing. Students also get the opportunity to attend the summer program in Mumbai and Berlin.
Location: Delhi, Mumbai, Berlin, London Fees: £500 Website: https://www.kcl.ac.uk/study/summer/spotlight/india-summer-programmes/indiasummer-programme
1 note
·
View note
Text
The Fallacious Commingling of Two Unrelated Hypotheses: “The Central Dogma” and “DNA Makes RNA Makes Protein”
Introduction
In the many dozens of undergraduate textbooks that have accumulated in my office in the last 15 years, the Central Dogma of Molecular Biology is almost invariably defined as the information-flow pathway from DNA to RNA to proteins. The wording is catchy: “DNA makes RNA makes protein,” and it is usually attributed to Francis Crick, but none of the books in my possession provides a traceable reference. In one case at least, the Central Dogma defined as “DNA makes RNA makes protein” is attributed to Watson and Crick (1953). In most of my textbooks it is also stated that the dogma was refuted by the discovery of process of reverse transcription from RNA to DNA by Howard Temin (1970) and independently by David Baltimore (1970).
One such example is shown below:

FIGURE 1. A typical description of the Central Dogma in a 2012 textbook. The gist of the description is that the dogma was put forward “in the early 1950s,” that the author of the dogma is Francis Crick, that the dogma postulates a “unidirectional flow of genetic information from DNA through RNA to protein, i.e., DNA makes RNA makes protein,” and that while the “broad thrust” of the dogma is correct, a number of modifications must be made to the basic scheme. Note, that no citation is given. Interestingly, the authors volunteer an explanation for the reason the Central Dogma was called “dogma,” rather than a “hypothesis,” “theorem,” or “conjecture.” According to these authors, Crick used the term “dogma” because "it was proposed without much evidence for the individual steps.”
Unfortunately, the Central Dogma has nothing to do with “DNA makes RNA makes protein,” and the catchy slogan has neither originated with Francis Crick, nor does it summarize (even approximately) the Central Dogma. For some reason, two independent hypotheses, the Central Dogma and the “DNA makes RNA makes protein” slogan, became mixed up with each other, hopelessly mangled, confounded, and confounding. In the following, I will try to make sense of this confusion.
The Historical Record of the “Central Dogma”
Francis Crick’s Central Dogma made its debut at the 1957 meeting of the Society for Experimental Biology. The paper based on his lecture appeared in 1958 in the journal Symposia of the Society for Experimental Biology (Crick 1958). In the paper, it is stated
“[The Central Dogma] states that once ‘information’ is passed into protein it cannot get out again. In more detail, the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information means here the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein.”
Because of the vagaries of the publication process in the pre-Internet era, the Central Dogma made its first appearance in print in 1957 in the popular science journal Scientific American (Crick 1957).
“This result illustrates very well a hypothesis which my colleagues and I call the Central Dogma: namely, that once information (meaning here the determination of a sequence of units) has been passed into a protein molecule, it cannot get out again, either to form a copy of the molecule or to affect the blueprint of a nucleic acid.”
In 1970, Temin and Mizutani (1970) and Baltimore (1970) independently announced their discovery of reverse transcription. Their papers in the journal Nature were accompanied by an unsigned News & Views item entitled “Central Dogma Reversed.” The anonymous author started the article as follows:
“The Central Dogma, enunciated by Crick in 1958 and the keystone of molecular biology ever since, is likely to prove a considerable oversimplification. That is the heretical but inescapable conclusion stemming from experiments done in the past few months in two laboratories in the United States. For the past twenty years the cardinal tenet of molecular biology has been that the flow or transcription of genetic information from DNA to messenger RNA and then its translation to protein is strictly one way. But on pages 1209 and 1211 of this issue of Nature, Baltimore and Mizutani and Temin claim independently that RNA tumor viruses contain an enzyme which uses the viral RNA as a template for the synthesis of DNA and thus reverses the direction of genetic transcription.”
The unsigned commentary ends with:
“[T]he Central Dogma, which like all dogmas has had a blinkering as well as an inspiring effect, [is] due for critical reappraisal.”
Crick was understandably miffed. His Central Dogma had nothing to do with directionality in the genetic-information flow. Baltimore, Mizutani, and Temin may have indeed refuted something out there in the literature, but it was certainly not his Central Dogma. Crick did not write “the transfer of information from DNA to RNA,” but “the transfer of information from nucleic acid to nucleic acid,” which of course does not preclude “the transfer of information from RNA to DNA.” Because the Central Dogma deals with the inability of genetic information to escape proteins, Crick, in effect anticipated the irrelevance of reverse transcription to his dogma. However, he felt the Nature News & Views piece was too influential to ignore, and here, again, after more than a dozen years since his original presentation, Crick was forced to repeat himself and explain the Central Dogma of Molecular Biology. This time, however, he abandoned his usual conciseness. No more succinct presentations in which the Central Dogma is presented by means of a couple of sentences or less. This time, he uses three pages, three figures, and sixteen chemical-pathway notations. The paper reads like a desperate attempt to cover all bases, leave nothing to the imagination, and prevent any misunderstanding in the future.
In his 1970 paper, Crick clearly formulates the Central Dogma in negative terms. There are three genetic information transfer pathways that can never occur: (1) Protein → Protein, (2) Protein → DNA, and (3) Protein → RNA. Everything else is OK. The Central Dogma can only be refuted if one of these three pathways is discovered in nature or in the laboratory.
The figures in Francis Crick’s 1970 article are particularly illuminating since it is clear that Crick as early as 1956 realized that the transfer of information from RNA to DNA is not a logical impossibility.
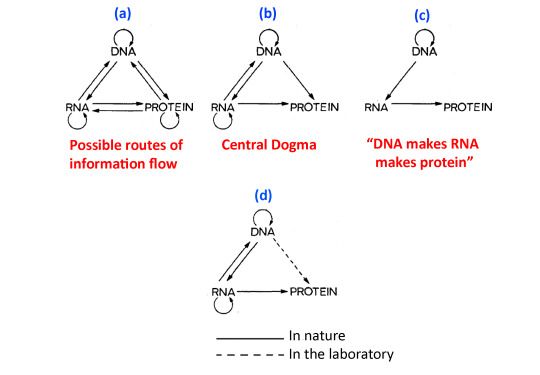
FIGURE 2. Information flow among DNA, RNA, and proteins. (a) In principle, there can be nine possible routes of information flow among DNA, RNA, and proteins. (b) The Central Dogma postulates that only six of these are allowable: DNA replication, RNA replication, transcription, reverse transcription, and translation. (c) One pathway of information flow, usually worded as “DNA makes RNA makes protein,” is frequently and incorrectly referred to as the Central Dogma. (d) The routes of information flow that are currently supported by experimental evidence either in nature or in the laboratory. Only one route of information flow that is allowable by the Central Dogma, i.e., from DNA to protein, has not been discovered in nature. The straight arrows represent a directional synthesis of a polymer on a different polymer template. The looped arrows represent self-template syntheses. Modified from Crick (1970) and Graur (2016).
By being exhaustively pedagogical, however, Crick (1970) penned one of his most repetitive and tedious articles I have read. The result, however, seems to me to be almost impervious to misunderstanding.
Sadly, it has been frequently and most probably deliberately misunderstood. In his Nobel lecture, Howard Temin (1975) declared:
“This transfer of information from the messenger molecule, RNA, to the genome molecule, DNA, apparently contradicted the “Central Dogma of Molecular Biology”, formulated in the late 1950’s.”
The use of the modifier “apparently” allowed Temin to distance himself from the strong claim of refutation and to soften the blow somewhat, but it is clear that his Central Dogma has nothing to do with Crick’s Central Dogma.
The Central Dogma continues to be confused with the slogan “DNA makes RNA makes protein” to this day. Take for an instance, a 2002 editorial entitled “Beyond the Central Dogma” in the journal Bioinformatics that was written by one of the top bioinformaticians in the world, Steven Henikoff (Henikoff 2002).
“The Central Dogma, ‘DNA makes RNA makes protein,’ has long been a staple of biology textbooks.”
“The Central Dogma was first challenged by the discovery of reverse transcription.”
Similarly, a certain J. Scott Turner, whose claim to fame is the assertion that evolutionary theory should not rely on genetics, declared in 2004 that “the Central Dogma is essentially dead” (Turner 2004).
Finally, a few months ago, a certain Anna Ritz from the Biology Department at Reed College wrote with absolute certainty that
“The phrase “DNA makes RNA makes protein,” while over-simplified, summarizes the transfer of information that has been collectively defined as the Central Dogma of molecular biology.”
“The Central Dogma is a concept coined by Francis Crick that, at its most fundamental level, describes the transfer of information within a cell (Crick 1970).”
Has this computer jock, who got her PhD from the ivy-league Brown University, even read any of Crick’s Central Dogma papers?
Why did Francis Crick Call His Hypothesis a “Dogma”?
In an interview with Horace Freeland Judson in 1975 (see Judson 1979; Thieffry and Sarkar 1998), Crick emphasized the original speculative dimension of the Central Dogma. He also feigned ignorance about the meaning of the word “dogma.”
“My mind was, that a dogma was an idea for which there was no reasonable evidence…”
“I just didn’t know what dogma meant. And I could just as well have called it the Central Hypothesis…”
“Dogma was just a catch phrase. And of course one has paid for this terribly, because people have resented the use of the term dogma, you see, and if it had been Central Hypothesis nobody would have turned a hair.”
I don’t buy this explanation; too coy, too contrived, too facetious. It is hard for me to believe that a person who grew up in a religious family and as a child attended church regularly would not be familiar with the term “dogma.” It is even harder for me to believe that a person who attended University College London and Cambridge University has never heard of “dogma.”
According to Sydney Brenner, the anti-religious Francis Crick used the ecclesiastical term “dogma” as a joke or a provocation (Ewing-Duncan 2006). I believe that this explanation is the correct one. My evidence is Francis Crick’s draft lecture from 1956 that is kept at the Wellcome Library in London (Crick 1956a).
The manuscript is entitled “Ideas on protein synthesis” and is dated October 1956. On the first page of the manuscript, two alternative appellations are considered: “The Doctrine of the Triad” and “The Central Dogma.” The two alternative names are dead giveaways. The 40-year-old Crick was obviously poking fun at Christianity, particular at the concept of the Trinity. In contrast, the 60-year-old Crick that was interviewed by Judson was a respectable sage and a Nobel laureate, who may not have wanted to disclose his youthful "indiscretions.”
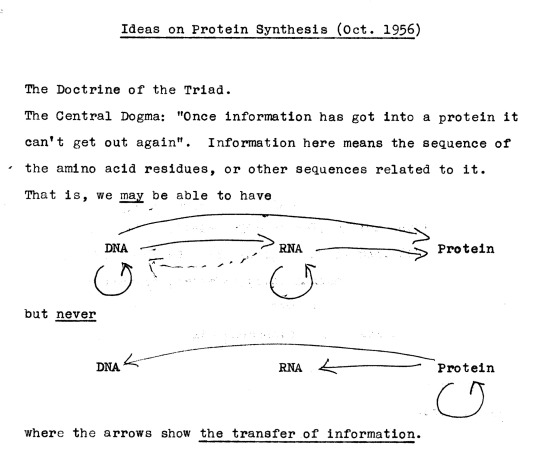
FIGURE 3. A draft manuscript for Crick’s 1957 lecture at the Meeting of the Society for Experimental Biology. The “Central Dogma” and the “Doctrine of the Triad” are obvious allusions to Church Dogma and the Doctrine of the Trinity.
The Meaning of “Information” in the Central Dogma
The term “information” is crucial to understanding the Central Dogma. According to Crick (1958), information is the sequence “either of bases in the nucleic acid or of amino acid residues in the protein.” Therefore, the Central Dogma makes three specific claims: (1) a sequence of amino acids cannot determine a sequence of ribonucleotides, (2) a sequence of amino acids cannot determine a sequence of deoxyribonucleotides, and (3) a sequence of amino acids cannot determine a sequence of amino acids (Morange 2008).
Crick was extremely adamant about the inability of a sequence of amino acids to determine a sequence of nucleotides for the simple reason that he could not envision a mechanism of reverse translation. In addition, Crick also noted that protein folding made the sequence of a protein inaccessible to any obvious means by which the sequence might be copied.
Some scientists, e.g., Jacques Monod (in Judson 1996), noted that the fact that it is impossible to transfer sequence information from protein to nucleic acid can be extended to mean that the phenotype cannot specifically alter the genotype and that the the soma cannot modify the germ line. Thus, the Central Dogma fitted perfectly well the Darwinian view (Morange 2008). Lamarckists of all shades and colors don’t like the Central Dogma very much (e.g., Noble 2013).
The Historical Record of “DNA Makes RNA Makes Protein”
From our modern vantage point it is difficult to grasp a time in which DNA, RNA, and proteins were considered to be independent molecules that had nothing to do with one another. Yet like everything else that is currently clear and obvious, we have to find out when did scientists realize that DNA serves as a template for making RNA and that proteins use RNA as a template?
Because of the chemical similarity between DNA and RNA, the idea that DNA makes RNA in the nucleus was formulated quite early (e.g., Caspersson 1941) and this seemed to have been accepted at least by some in the scientific community. The idea that RNA makes protein in the cytoplasm (e.g., Boivin 1949; Dounce 1952) was much less intuitive and took longer to be accepted (Brachet 1942, 1954; Caspersson 1947; Boivin et al. 1949; Dounce 1953).
The first clear enunciation of the connection between DNA, RNA, and proteins and the unidirectionality of transcription and translation is found in an article by André Boivin and Roger Venderly (1947):
“[A] great number of different desoxyribonucleic and ribonucleic acids exist in each cell: desoxyribonucleic acids in the nucleus (genes) and ribonucleic acids in the cytoplasm (microsomes). Through catalytic actions the macromolecular desoxyribonucleic acids govern the building of macromolecular ribonucleic acids, and, in turn, these control the production of cytoplasmic enzymes.”
We now take for granted that we have immediate access to the literature, regardless of the publication venue and the nationality of the authors. We also live in a world in which the impact of articles is immediate. This was not always so in the pre-Googlian world. Each scientist in these “ancient” times used to have a restricted list of journals that they consulted, and each scientist had a limited circle of colleagues with which they corresponded. Nowadays, in between press releases, Twitter, email, and online journals, I, for instance, can discover in a few seconds who said what and where on my favorite topic, which happens to be pseudogenes (dead genes). With Google Translate it doesn’t even matter much if the article is written in French or Russian, although, of course, nowadays the vast majority of article that are worth reading are in English.
In the 1940s and 1950s, the situation was different; scientific findings took time to percolate the consciousness of the scientific community. And many important findings were simply forgotten, waiting for others to reinvent the wheel. Unfortunately, for Boivin and Venderly (1947), articles written in French became unfashionable in the American-dominated post World War II science.
Take for example Erwin Chargaff, who seemed to have been completely oblivious of Cassperson, Boivin, Dounce, Brachet, and Venderly, when in 1954 he expressed the following opinion at a meeting dealing with radiation effects on cells (Sherman 1954).
“I seem to gather that the theory now is that DNA makes RNA and RNA makes protein. This may be so in special cases. I think there is some evidence that DNA makes DNA and RNA makes RNA. In fact, there is little chemical relationship [ ] between the total DNA of the cell and the RNA. We have looked for this but there does not seem to be any.”
This is startling; Chargaff essentially denies that DNA makes RNA and RNA makes protein. For him the fact that the amounts of RNA and DNA in a cell are not positively correlated with each other constitutes a refutation of this particular theory. He could only bring himself to admit two things: (1) that “DNA makes DNA” and “RNA makes RNA,” and (2) that “DNA makes RNA and RNA makes protein” may be true in “special cases.”
Chargaff’s comment, however, teaches us an important historical lesson, i.e., that a slogan resembling “DNA makes RNA makes protein” was known at least three years before Francis Crick came up with the Central Dogma.
The first two instances that the exact slogan “DNA makes RNA makes protein” is recorded in the literature are in Tweet (1961) and Tyler (1963):
“M. B. Hoagland (Harvard Medical School) next discussed the genetic code and protein synthesis. The dogma that DNA makes RNA makes protein is by now as thoroughly ingrained in all readers of the Scientific American as is the doctrine of the triplet code.”
and
“I wish only to remark that the clonal selection theories do not eliminate the assumption of template mechanisms, but simply restrict these to the 'DNA makes RNA makes protein' part of the process.”
In both cases, the slogan is mentioned in passing as something familiar to the “readers of the Scientific American,” so although I have failed to find pre-1961 occurrences, we can be certain that antecedents of the” DNA makes RNA makes protein” slogan exist.
According to Dawkins (2004), “DNA makes RNA makes protein” sounds pithy and clever, but “it is too pithy and not clever enough.” It is merely a summary of research findings, rather a theoretical principle like Crick’s Central Dogma.
The popularity of the catchy slogan “DNA makes RNA makes protein” has increased continuously since its inception. Sydney Brenner, for instance, liked the slogan so much that he extended it to “DNA makes RNA makes protein makes money” (Ewing-Duncan 2006).
It is important to note, that the principle summarized by the slogan “DNA makes RNA makes protein,” which is usually referred to by the erroneous moniker the Central Dogma, had already been given a name in 1953. It was called the Template Hypothesis (Dounce 1953):
“There can be no objection at the present time to assuming that the genes are the templates, but it is not necessary to assume that the genes act directly as templates for protein synthesis. If we accept the suggestion by Mazia (1952) that genes are composed of deoxyribonucleic acid, then it conceivably happen that the deoxyribonucleic acid gene molecules would act as templates for ribonucleic acids synthesis, and that the ribonucleic acids synthesized on the gene templates would in turn become templates for protein synthesis in the nucleus or cytoplasm or both.”
In a textbook from 1960 entitled The Biological Role of Nucleic Acids, we find a clear explanation of the Template Hypothesis (Brachet 1960):
“The results obtained from experiments on unicellular organisms are in good agreement with views which have been expressed repeatedly since Caspersson (1941, 1950) presented them first. DNA, which is the primary genetic substance, would synthesize RNA; proteins would, in turn, be synthesized under the influence of RNA. The Template Hypothesis provides an easy explanation for specificity. Specific DNA molecules (or parts of molecules) corresponding to each gene would act as a template for RNA; there would thus be as many specific RNA molecules as there are genes. Finally, each of these specific RNA molecules would act as a template for a specific protein, according to the mechanism discussed in Chapter 2. Such a scheme corresponds to the now familiar slogan: DNA makes RNA, and RNA makes protein.”
The term Template Hypothesis enjoyed some popularity until the middle 1960s (e.g., Landman and Spiegelman 1955; Berg and Offengand 1958; Siekevitz 1959; Speyer 1965). After 1965 and the publication of Watson’s textbook, the term fell into disuse, and by 1970 it disappeared completely from the scientific literature.
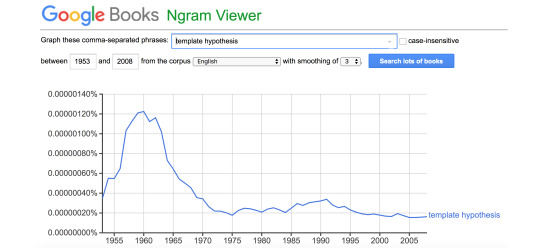
FIGURE 4. The rise and fall of the term “Template Hypothesis” from the literature.
Who Should We Blame for the Confusion?
Here we attempt to find out who equated the Central Dogma with “DNA makes RNA makes protein,” and who attributed this mishmash to Francis Crick.
The first part of the query has an easy answer; the universally recognized villain is James Watson. In Chapter 10 of his influential textbook, Watson (1965) dedicates an entire section to the Central Dogma. In it he writes:
“We should first look at the evidence that DNA itself is not the direct template that orders amino acid sequences. Instead the genetic information of DNA is transferred to another class of molecules, which then serve as the protein template. These intermediate templates are molecules of ribonucleic acid (RNA), large polymeric molecules chemically similar to DNA. Their relation to DNA and protein is summarized by the [following] formula (often called the Central Dogma)
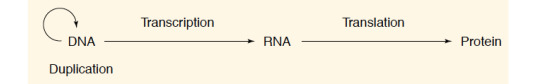
where the arrows indicate the direction of transfer of the genetic information. The arrow encircling DNA signifies that it is the template for self-replication; the arrow between DNA and RNA indicates that all cellular RNA molecules are determined by DNA templates. Correspondingly, all protein sequences are determined by RNA templates. Most importantly, both these latter arrows are unidirectional, that is, RNA sequences are never copied on protein templates; likewise never acts as a template for DNA.”
In the Summary of Chapter 10, Watson adds one additional attribute to the Central Dogma:
“In a given gene only one of the two DNA strands is copied. The molecular basis of this differential transcription is not known.”
Finally, in the Glossary, we find:
Central Dogma. The basic relationship between DNA, RNA, and protein: DNA serves as a template for both its own duplication and the synthesis of RNA; and RNA, in turn, is the template in protein synthesis.
We must first emphasize that the Central Dogma in James Watson’s textbook is unattributed. Mercifully, he does not associate this atrocity to Francis Crick. Indeed, in his autobiographical gossipy book The Double Helix (Watson 1968), he attributes the Central Dogma to himself. The second thing we must emphasize is that the slogan “DNA makes RNA makes protein” is nowhere to be found. We must deduce it from the figure, where the arrows stand for “makes.” Of course, the flow of genetic information in Watson’s figure should be literally translated into “DNA makes DNA makes RNA makes protein.”
There are four main points in James Watson’s description of the Central Dogma that are different from Francis Crick’s original phrasing.
First, by stating that “all cellular RNA molecules are determined by DNA templates,” Watson denies the possibility of RNA replication, which is perfectly OK in Crick’s version.
Second, by emphasizing the unidirectionality of the information transfer, e.g., “RNA never acts as a template for DNA,” Watson denies the possibility of reverse transcription, which Crick never did
Third, Watson does not believe that DNA can serve as template for translation into proteins without an RNA intermediary, which Crick believed, at least in principle, to be possible.
Finally, Watson’s Central Dogma declares that only one strand of the gene is transcribed into RNA, a position on which Crick’s dogma has absolutely no opinions.
As we shall see in the next section, all the new attributes that Watson piled on top of Crick’s Central Dogma have been refuted.
At this point we should note (at least parenthetically) that before 1965, writers other than Watson referred to the slogan “DNA makes RNA makes protein” as a “dogma,” but never as a Central Dogma. One such example was already mentioned above (Tweet 1963).
Because Watson’s fake “Central Dogma” appeared in an extremely popular textbook—until 2013, it went through eight editions while accumulating five coauthors—it gradually eclipsed Crick’s original Central Dogma and gained widespread currency as expressing “a truth beyond doubt” (Allchin 2017). After the publication of his textbook, the term Template Hypothesis completely disappeared to be replaced by the Central Dogma (Figure 4).
While James Watson is singularly to blame for the confusion between the Central Dogma and “DNA makes DNA makes RNA makes protein,” it is very difficult to find out exactly who added Francis Crick’s name to the unholy mixture. What is certain is that by the late 1970s and early 1980s it has become common to add Crick's name to Watson’s fake “Central Dogma.” In particular, textbooks (e.g., Tribe et al. 1978) relished in attributing a great phrase to a great man:
“One of the main aims of the book was to examine the evidence that DNA is the basic material of heredity, and that ‘DNA makes RNA makes protein’—the ‘Central Dogma’ of biology as postulated by Crick.”
Watson’s fake “Central Dogma” as a Convenient Straw Man
A straw man is a farfetched argument set up for the sole purpose of being easily refuted. Straw-man statements are usually caricatures of important scientific theories or postulates that are erected for the purpose of claiming a victory against eminent scientists, such as Crick, Darwin and Einstein. By claiming that Watson’s distortion of the Central Dogma is the same as Crick’s Central Dogma, a person can “score” a victory against one of the most respectable scientists of the twentieth century. In a similar manner, creationists can claim victory over Darwin by refuting something they claim is junk DNA, but isn’t (Graur et al. 2013).
All the four elements that Watson added to the Central Dogma have been refuted.
The first to be refuted was the claim that RNA replication is impossible. The discovery of RNA-dependent RNA replication by Sol Spiegelman and his colleagues (Haruna et al. 1963; Doi and Spiegelman; Spiegelman 1963) preceded Watson’s fake dogma by two years. We also know for certain that Sol Spiegelman was aware of Crick’s Dogma, most probably because in 1956, Crick wrote a letter to Spiegelman to which the Central Dogma draft in Figure 3 was attached (Crick 1956b).
The second to be refuted was the claim that reverse transcription is impossible (Anonymous 1970), which has already been dealt with in the Introduction.
Subsequently, the claim that only one strand of the gene is transcribed into RNA was refuted by empirical findings in viruses (Barrel et al. 1976), bacteria (Smith and Parkinson 1980), and animals (Spencer et al. 1986).
The most important difference between Watson’s faux Central Dogma and Crick’s real Central Dogma is the claim that DNA cannot be translated directly into proteins. This topic deserves to be discussed separately.
Can Protein be Synthesized Directly on a DNA Template?
Francis Crick’s Central Dogma does not preclude the synthesis of proteins in the absence of RNA. So far, such a process has never been found in nature, and chances are it never will. But, is such a process possible?
McCarthy and Holland (1965) demonstrated that aminoglycoside antibiotics, e.g., streptomycin and neomycin, can potentiate the activity of single stranded DNA from Escherichia coli as a template for protein synthesis in the absence of mRNA. The reason for the need of aminoglycoside antibiotics is that that one step in the translation process depends on the ribosome forming bonds with the two OH groups of the nucleotides at all three positions of the codon. If the codons are made of DNA (deoxycodons), the translation process leads to either incorrect tRNA selection or no tRNA selection. The presence of an aminoglycoside antibiotic allows the bonding between ribosome and anticodons and hence, translation, to proceed even in the absence of OH groups (Ogle and Ramakrishnan 2005).
Similar results were obtained by Uzawa et al. (2002) in the extreme thermophilic bacterium Thermus thermophilus and in the hyperthermophilic acidophilic archaebacterium Sulfolobus tokodaii.
There are two possible reasons why DNA-dependent translation does not exist in nature. The first is that there are insurmountable functional issues that prevent the translation of proteins directly from single stranded DNA from realizing itself. In other words, the process biochemically unfeasible. The laboratory results described above as well as many other reports in the literature lead me to believe that this is not the true reason.
I suspect the most parsimonious reason is that DNA simply was not around yet by the time translation evolved and was optimized my natural selection. This claim is intimately related to the concept of the RNA world.
The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of proteins and DNA. .
Alexander Rich first proposed the concept of the RNA world in 1962. The term RNA world was coined 24 years later by Walter Gilbert (1986). Currently, the evidence for an RNA world is considered strong enough that the hypothesis has gained wide acceptance (Neveu et al. 2013; Vázquez-Salazar and Lazcano 2018).
The first stage in the RNA world is postulated to involve self replicating RNAs. Next, proteins were added, most probably as components of ribonucleoproteins (RNPs). During this stage, sometimes referred to as the RNP world, the genetic code and translation evolved. The end of the RNA/RNP world came with the emergence of DNA and long proteins (Cech 2012). The emergence of DNA and enzymes had nothing to do with translation. DNA has better stability and durability than RNA, which may explain why it became the predominant information storage molecule, and protein enzymes may have come to replace RNA-based ribozymes as biocatalysts because their greater abundance and diversity of their monomeric components (20 amino acids versus 4 nucleotides).
In evolution it is very difficult to improve on a system that works, especially if the improvement requires many steps, and if the intermediate steps (between the system that works and the system that would work better) are deleterious.
Because of historical contingency, “RNA makes protein” most probably represents an achievable solution, rather than an optimal solution.
Silly Fake Faux Central Dogmas
In addition to Watson’s Central Dogma, the scientific and popular literature abounds in silly definitions of the “Central Dogma,” that are promptly and invariably refuted by those who invented these dogmas, usually to the sound of self-righteous chest thumping and proclamations of superiority over the rest of the scientific world.
In his Nobel Lecture, Andrew Fire (2006) proclaimed that the Central Dogma is dead. His definition of the Central Dogma was summarized as “double stranded DNA makes single stranded RNA makes protein.” Not surprisingly, his RNA interference (RNAi), which involves double stranded RNA, was claimed to refute the Central Dogma.
Even ribozymes have been used to “refute” the Central Dogma. Take for example, Nils Walter and David Engelke (2002):
“Twenty years ago, it became clear that ribonucleic acids, or RNAs, are used as catalysts in living cells, in addition to their known roles in information storage and as molecular architectural frameworks. This idea was so profoundly contrary to the Central Dogma of molecular biology that it resulted in the award of a Nobel prize to two of the early proponents, Thomas Cech and Sidney Altman.”
In Walter and Engelke’s version of the Central Dogma, RNAs are not supposed to have catalytic activity, which, of course, is a claim that was made by neither Crick nor Watson.
In 2015, two punctilious ignoramuses, Manel Esteller and Sonia Guil, published a paper in Trends in Biochemical Sciences in which they decided to invent ye another definition of the Central Dogma. In their Central Dogma, “one gene gives rise to one RNA to produce one protein,” which seems to be an extension of a suggestion originally proposed by Lucien Cuénot (1903) and elaborated upon by George Beadle and Edward Tatum (1951). This hypothesis was referred to as the one gene-one enzyme hypothesis (Luria 1947; Horowitz 1948) and the one gene-one protein hypothesis (Beadle 1948). As expected, Esteller and Guil (2015) thoroughly and totally refute this “new and improved” Central Dogma. Where did these people even come up with this definition?
Denis Noble, who is a Commander of the Most Excellent Order of the British Empire, Fellow of the Royal Society, Fellow of the Royal College of Physicians of London, Fellow of the Academy of Medical Sciences, Professor Emeritus, and co-Director of Computational Physiology at Oxford, “refuted” not only the Central Dogma, but also Darwinism and all “the foundations of evolutionary biology” (Noble 2013). How did it do that? By claiming that the Central Dogma postulates that “genomes are isolated from the organism and the environment.”
Even psychologists have got into the game of refuting the fake faux Central Dogma. Gilbert Gottlieb (1998), for instance, used the following definition:
"The Central Dogma of Molecular Biology holds that ‘information' flows from the genes to the structure of the proteins.”
By completely ignoring Crick’s definition of “information,” Gottlieb manages to turn any influence of one entity upon another into a transfer of information. For example, he claims that Crick’s injunction against protein → DNA information transfer has been falsified by the fact that some proteins can bind DNA and affect transcription:
Regarding Protein → DNA transfer, there has long been recognized a class of regulative proteins that bind to DNA, serving to activate or inhibit DNA expression (i.e., turning genes on or off).
By similarly ignoring Crick’s definition of “information,” Gottlieb also claims that the injunction against protein → protein information transfer has also been rejected by the discovery of prions—a subject that will be dealt with in detail in the next section.
Gottlieb’s silly definitions of information and information transfer can lead to some interesting observation. For example, if I were to punch Gottlieb in the face—something that I cannot do currently because the guy died in 2006—this would constitute a flow of information. I can only imagine standing before a judge and accused of unlawful information transfer and aggravated battery.
Finally there are those who want to one-up Watson. Pedro Romero and colleagues came up with this definition of the central dogma (Romero et al. 2000):
“Indeed, the current Central Dogma of Molecular Biology states that information flows in the following manner: DNA → mRNA → amino acid sequence → 3D structure → function...”
I was left wondering why Romero et al. decided to stop at function and not continue with ...function → phenotype → life → the universe → everything. (I dearly hope that from the great beyond, Douglas Adams will forgive my appropriation.)
It is, of course, very easy to define the Central Dogma in a manner that can never be refuted. It seems also quite easy to get away with attributing stuff to Francis Crick. It has been done so many times; who would know?
Central Dogma: The claim the Donald Trump is a thieving thug in the service of Vladimir Putin and Russian oligarchs.
Prions: The Only Attempt at Refuting Crick’s Central Dogma Rather than Watson’s Central Dogma
Prions are infectious protein particles that can cause such degenerative diseases as scrapie in sheep and goats and transmissible spongiform encephalopathy (TSE)—popularly known as Mad Cow Disease—in humans.
Stanley Prusiner, the scientist who discovered prions, was fascinated by questions concerning the mode of prion replication. The particles seems devoid of both DNA and RNA, so how do they replicate? In his seminal paper in Science, Prusiner (1982) raised five possibilities:
The first two possibilities envisioned the existence of “undetected nucleic acids” within the prions: (1) the undetected nucleic acids code for the prion protein or proteins, or (2) the undetected nucleic acids activate transcription of host genes coding for prion protein or proteins.
The other three possibilities assumed that prions do not contain nucleic acids: (3) prions activate transcription of host genes coding for the prion protein or proteins, (4) prions code for their own replication by reverse translation, or (5) prions code for their own replication by protein-directed protein synthesis.
Prusiner correctly recognized that the Central Dogma would be “contradicted” if prions use methods (4) or (5), i.e., if they code for their own biosynthesis either through reverse translation or protein-directed protein synthesis.
As it turned out none of Prusiner’s five hypotheses turned out to be true. The mode of prion replication is unique.
Prions turned out to be run-of-the mill proteins encoded by a run-of-the mill gene. In humans, the PrP protein is encoded by the two-exon PRNP gene, which is located on the short arm of chromosome 20. The protein that prions are made of, PrP, is found throughout the body, even in healthy people and animals. In humans, PrP is a normal nonessential mammalian cell surface protein of uncertain function. PrP found in infectious material, on the other hand, has a different three-dimensional (3D) structure and is resistant to proteases, the enzymes in the body that can normally break down proteins. The normal form of the protein is called PrP-C, while the infectious form is called PrP-Sc—the C refers to “cellular,” while the Sc refers to “scrapie,” the prototypic prion disease.
PrP-C is found on the membranes of cells. It has 209 amino acids (in humans), one disulfide bond, a molecular mass of 35–36 kDa, and a mainly alpha-helical structure. The normal protein is not sedimentable; meaning that it cannot be separated by centrifuging techniques. PrP-C binds copper ions with high affinity, is readily digested by proteinase K, and can be liberated from the cell surface in vitro by the enzyme phosphoinositide phospholipase C. Sadly, the function of the normal protein is unknown.
The infectious isoform of PrP, known as PrP-Sc, is able to convert normal PrP-C proteins into the infectious isoform by changing their their 3D shape, which, in turn, will alter the way the proteins interconnect. PrP-Sc always causes prion disease. Aggregations of PrP-Sc isoforms form highly structured amyloid fibers, which accumulate to form plaques. It is unclear whether these aggregates are the cause of cell damage or are simply a side-effect of the underlying disease process. The end of each fiber acts as a template onto which free protein molecules may attach, allowing the fiber to grow. Under most circumstances, only PrP-Sc molecules with an identical amino acid sequence to the infectious PrP-Sc are incorporated into the growing fiber. However, rare cross-species transmission is known.
What is interesting to note is that the amino acid sequence of the PrP-C protein and that of the PrP-Sc protein are identical. The only difference between the proteins is their 3D structure.
So can prions be construed as a refutation of Crick’s Central Dogma? Does this structural “contagiousness” contradicts the Central Dogma? The answer is a big fat No! Francis Crick defined “information” as “the sequence of amino residues in a protein or the sequence of nucleotides in DNA or RNA.” According to this definition, a prion does not transmit information from protein to protein.
Is the Central Dogma Alive? Dead? In Schrödinger’s Cat’s Limbo
In many respects the Central Dogma is a Rorschach Blot onto which scientists, philosophers, fiction and nonfiction writers project their concerns, their prejudices, their peeves and their likes with no hesitation over altering the dogma to meet their distinctive needs. In the literature, one can find literally thousands of refutations, yet each of those defines the Central Dogma in ways that are at variance with the original definiton by Francis Crick.
So is the Central Dogma alive? Paraphrasing Mark Twain's frequently misquoted quip, reports of the death of the Central Dogma has been grossly exaggerated. Richard Dawkins (2004) put it best when he wrote:
“[T]he Central Dogma has never been violated and my bet is that it never will. The genetic code, whereby nucleotide sequences are translated into amino acid sequences, is irreversible.”
Not only is the Central Dogma of Molecular Biology alive and well, the phrase has proved so attractive that we now have the Central Dogma of Neurology (which has probably reached the end of its usefulness), the Central Dogma of Palliative Care (which needs rethinking), the Central Dogma of Flow Cytometry (which may have lost some of its validity by the finding that mammalian cells are much less refractive than expected), and many, many more.
Additional Reading Suggestions
For a comprehensive discussion of the history of prions, see Reeves (2002). An earlier discussion on the Central Dogma can be found in Moran (2007).
References
Allchin D. 2017. Sacred Bovines: The Ironies of Misplaced Assumptions in Biology. Oxford University Press, New York
Anonymous. 1970. Nature. Central dogma reversed. 226:1198–1199.
Baltimore D. 1970. RNA-dependent DNA polymerase in visions of RNA tumor viruses. Nature 226:1209–1211.
Barrel BG, Air GM, Hutchison CA. 1976. Overlapping genes in bacteriophage ΦX174. Nature 264:34-40.
Beadle GW. 1948. Some recent developments in clinical genetics. Prog. Chem. Org. Nat. Prod. 5:300–310.
Beadle GW, Tatum EL. 1941. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. USA 27:499–506.
Berg P, Offengand EJ. 1958. An enzymatic mechanism for linking amino acids to RNA. Proc. Natl. Acad. Sci. USA 44:78–86.
Boivin A, Vendrely R. 1947. Sur le rôle possible des deux acides nucléiques dans la cellule vivante. Experientia 3:32–34.
Boivin A, Vendrely R, Tulasne R. 1949. La spécificité des acides nucléiques chez les êtres vivants, spécialement chez les Bactéries. Coll. Inter. CNRS. 8:67–78.
Brachet J. 1942. La localisation des acides pentose nucléiques dans les tissus animaux et les oeufs d’amphibiens en voie de développement. Archi. Biol. 53:207–257.
Brachet J. 1954. Nuclear control of enzymatic activities. pp. 91–104. In: Kitching JA (ed). Recent Developments in Cell Physiology. Butterworths Scientific Publications, London.
Brachet J. 1960. The Biological Role of Nucleic Acids. Elsevier, Amsterdam.
Caspersson T. 1941. Studien über den Eiweißumsatz der Zelle. Naturwissenschaften 29:33–48.
Caspersson T. 1947. The relations between nucleic acid and protein synthesis. Symp. Soc. Exp. Biol. 1:127–151.
Cech TR. 2012. The RNA worlds in context. Cold Spring Harbor Perspect. Biol. 4:a006742.
Cobb M. 2017. 60 years ago: Francis Crick changed the logic of biology. PLoS Biol. 15:e2003243.
Crick FHC. 1956a. Ideas on protein synthesis. http://profiles.nlm.nih.gov/SC/B/B/F/T/_/scbbft.pdf.
Crick FHC. 1956b. Letter from Francis Crick to Sol Spiegelman. https://profiles.nlm.nih.gov/ps/retrieve/ResourceMetadata/PXBBDP.
Crick FHC. 1957. Nucleic acids. Sci. Am. 197(3):188–203.
Crick FHC. 1958. On protein synthesis. Symposia of the Society for Experimental Biology 12:138–163.
Crick FHC. 1970. Central dogma of molecular biology. Nature 227: 561–563.
Cuénot L. 1903. L’hérédité de la pigmentation chez les souris. Arch. Zool. Exp. Gen. 4: 33–41.
Dawkins R. 2004. Extended phenotype—but not too extended. A Reply to Laland, Turner and Jablonka. Biol. Phil. 19:377–396.
Doi RH, Spiegelman S. 1963. Conservation of a viral RNA genome during replication and translation. Proc. Natl. Acad. Sci. USA 49:353–360.
Dounce AL. 1952. Duplicating mechanism for peptide chain and nucleic acid synthesis. Enzymologia 15:251–258.
Dounce A. 1953. Nucleic acid template hypothesis. Nature 172:541–542.
Ewing-Duncan D. 2006. Masterminds: Genius, DNA, and the Quest to Rewrite Life. Harper Perennial, New York.
Fire AZ. 2006. Nobel Lecture: Gene silencing by double stranded RNA. https://assets.nobelprize.org/uploads/2018/06/fire_lecture.pdf?_ga=2.186251562.241175507.1535556506-866357347.1535358041
Gilbert W. 1986. The RNA world. Nature 319: 618.
Gottlieb G. 1998. Normally occurring environmental and behavioral influences on gene activity: From central dogma to probabilistic epigenesis. Psych. Rev. 105:792–802.
Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. 2013. On the immortality of television sets: "function" in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol. 5:578–590.
Graur D. 2016. Molecular and Genome Evolution. Sinauer, Sunderland, Massachusetts.
Guil S, Esteller M. 2015. RNA–RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem. Sci. 40:248–256.
Haruna I, Nozu K, Ohtaka Y, Spiegelman S. 1963. An RNA "replicase" induced by and selective for a viral RNA: Isolation and properties. Proc. Natl. Acad. Sci. USA 50:905–911.
Henikoff S. 2002. Beyond the central dogma. Bioinformatics 18:223–225.
Holland JJ, McCarthy BJ. 1964. Stimulation of protein synthesis in vitro by denatured DNA. Proc. Natl. Acad. Sci. USA 52:1554–1561.
Horowitz NH. 1948. The one gene-one enzyme hypothesis. Genetics 33:612.
Judson HF. 1979. The Eighth Day of Creation. Simon & Schuster, New York.
Koonin EV. 2012. Does the central dogma still stand? Biol. Direct 7:27.
Landman OE, Spiegelman S. 1955. Enzyme formation in protoplasts of Bacillus megaterium. Proc. Natl. Acad. Sci. USA 41:698–704.
Luria SE. 1947. Recent advances in bacterial genetics. Bacteriol. Rev. 11:1–40.
Mazia D. 1952. Physiology of the cell nucleus. pp. 77–122. In Guzmán Barrón ES (ed) Modern Trends in Physiology and Biochemistry, Academic Press, New York.
McCarthy BJ, Holland JJ. 1965. Denatured DNA as a direct template for in vitro protein synthesis. Proc. Natl. Acad. Sci. USA 54:880–886.
Moran L. 2007. Basic concepts: The central dogma of molecular biology. http://sandwalk.blogspot.com/2007/01/central-dogma-of-molecular-biology.html.
Morange. 2008. What history tells us. XIII. Fifty years of the Central Dogma. J. Biosci. 33:171–175.
Neveu M, Kim HJ, Benner SA. 2013. The "strong" RNA world hypothesis: Fifty years old. Astrobiology 13:391–403.
Noble D. 2013. Physiology is rocking the foundations of evolutionary biology. Exp. Physiol. 98:1235–1243.
Ogle JM, Ramakrishnan V. 2005. Structural insights into translational fidelity. Annu. Rev. Biochem. 74:129–177.
Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136–144.
Reeves C. 2002. An orthodox heresy: Scientific rhetoric and the science of prions. Sci. Commun. 24:98–122
Rich A. 1962. On the problems of evolution and biochemical information transfer. pp 103–126. In: Kasha M, Pullman B (eds). Horizons In Biochemistry, Academic Press, New York.
Ritz A. 2018. Programming the central dogma: An integrated unit on computer science and molecular biology concepts. pp. 239–244. In: Association for Computing Machinery Special Interest Group in Computer Science Education. ACM, Baltimore, Maryland.
Romero P, Obradovic Z, Dunker AK. 2000. Intelligent data analysis for protein disorder prediction. Artificial Intel. Rev. 14:447–484. Sherman FG. 1954. Cellular biochemistry. pp. 54–85. In: Patt HM (ed) Basic Mechanisms in Radiobiology. III. Biochemical Aspects. National Academy of Science National Research Council, Washington DC.
Siekevitz P. 1959. The cytological basis of protein synthesis. Exp. Cell Res. 7S:90–110.
Smith RA, Parkinson JS. 1980. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. USA 77:5370-5374.
Spencer CA, Gietz RD, Hodgetts RB. 1986. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature 322:279–281,
Speyer JF. 1965. Mutagenic DNA polymerase. Biochem. Biophys. Res. Commun. 21:6-8.
Spiegelman S. 1963. Information transfer from the genome. Fed Proc. 22:36–54.
Temin HM, Mizutani S. 1970. RNA dependent DNA polymerase in the visions of Rous sarcoma virus. Nature 226:1211–1213.
Temin HM. 1975. Nobel Lecture: The DNA provirus hypothesis. The establishment and Implications of RNA-directed DNA synthesis. https://assets.nobelprize.org/uploads/2018/06/temin-lecture.pdf?_ga=2.147971252.241175507.1535556506-866357347.1535358041.
Thieffry D, Sarkar S. 1998. Forty years under the central dogma. Trends Biochem Sci. 23:312–316.
Tribe MA, Tallan I, Eraut MR. 1978. Basic Biology Course, Unit 5, Volume 12, Case Studies in Genetics: Aspects of Heredity. Oxford University Press, Oxford
Turner JS. 2004. Extended phenotypes and extended organisms. Biol. Phil. 19:327–352.
Tweet AG. 1963. Perspectives on recent progress in biophysics. Physics Today 16(12):34-38.
Tyler A. 1963. The manipulations of macromolecular substances during fertilization and early development of animal eggs. Am. Zool. 3:109–126.
Uzawa T, Yamagishi A, Oshima T. 2002. Polypeptide synthesis directed by DNA as a messenger in cell-free polypeptide synthesis by extreme termophiles, Thermus thermophilus HB27 and Sulfolobus tokodaii strain 7. J. Biochem. 131:849–853.
Vázquez-Salazar A, Lazcano A. 2018. Early life: Embracing the RNA world. Curr. Biol. 28:1166–1167.
Walter NG, Engelke DR. 2002. Ribozymes: Catalytic RNAs that cut things, make things, and do odd and useful jobs. Biologist 49:199–203.
Watson JD. 1965. Molecular Biology of the Gene. W. A. Benjamin, New York.
Watson JD, Crick FHC. 1953. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 171:737–738.
4 notes
·
View notes
Text
Science and Chemistry Classes
As a postdoctoral research associate in the lab of BTI faculty member Frank Schroeder, Max Helf saw his labmates continually struggle when they were analyzing data. So, he decided to do something about it and developed a free, open-source app called Metaboseek, which is now essential to the lab's work.
The Schroeder lab studies the roundworm Caenorhabditis elegans, one of the most successful model systems for human biology, to discover new metabolites that govern evolutionarily conserved signaling pathways and could be useful as leads for the development of new pharmaceuticals or agrochemicals. The researchers accomplish this task by comparing the metabolites between two different worm populations—a process called comparative metabolomics.
Given that samples routinely have more than 100,000 compounds in them, computational approaches are essential to perform the analysis.
The team had been relying on software packages that did not offer the required level of flexibility to easily customize analysis parameters. That limitation, and the lack of a suitable graphical user interface, meant Helf's colleagues faced the cumbersome task of visually inspecting mounds of data—for example, to spot possible false positives—and jumping between several other software tools to confirm and filter out those meaningless results.
"It just seemed very inefficient to me, and I couldn't get over the shortcomings of other software solutions for this problem," Helf said. "I thought there had to be an easier way, so I started to write code for my own software."
Helf developed the initial version of his software in 2017, and continued to improve it over the next two years. "Besides addressing the problems my labmates were already facing, I talked to them about what else held them back—what they wanted to do but weren't even trying—and built those features in the app," said Helf, who is now a bioinformatics product manager at proteomics company Biognosys AG. "I wanted this new tool to be user-friendly and accessible to anyone who does chemical biology."
The result was Metaboseek, an app with a graphical interface that incorporates multiple data analysis tools that non-coding researchers would otherwise not have. The app streamlines the analysis of comparative metabolomics data by helping the researcher determine which data features are real and letting them dig deeper into those features—all within the same tool.
"Max did this without me even requesting it," Schroeder said. "Before I knew that this was happening, there was Metaboseek. We started using it, and now our lab and many collaborators couldn't exist without it."
In a study published in Nature Communications on February 10, Schroeder's team provided proof-of-concept for Metaboseek by applying it to an important fat metabolism pathway that hadn't yet been studied: the α-oxidation pathway in C. elegans that helps break down a class of fatty acids.
Using Metaboseek, the team found that roundworms lacking a key gene in the α-oxidation pathway accumulated hundreds of previously unreported metabolites. The findings are important because α-oxidation is a basic biochemical pathway in worms that is conserved in humans, Schroeder said.
"Bennett Fox did the chemistry work, so this study was a nice collaboration between the two postdocs," added Schroeder, who is also professor in Cornell University's Department of Chemistry and Chemical Biology.
According to Schroeder and Helf, there are a few reasons why there aren't a lot of good analytic tools for comparing metabolomics data. First, comparative metabolomics is a relatively young field compared with other data-heavy fields of biology like genomics (which focuses on DNA) and proteomics (which focuses on proteins), so there hasn't been enough time to develop software tools and database infrastructure.
Additionally, over the last decade, the advent of affordable, ultra-high-resolution mass spectrometers for collecting metabolomics data has increased by perhaps more than tenfold the amount of data one sample can generate—creating an even greater need for sophisticated tools that can keep up with the flood of data.
Metaboseek meets these needs with an array of features for analyzing various types of data to aid compound identification, structure determination, assignment of metabolites to families based on structural similarities, tracking radiolabeled compounds, and more.
0 notes
Text
June 28, 2021
Well, it's been a few years since I wrote anything here. Mostly because these past few years have been an absolute whirlwind. My last post ended with Sean and I almost becoming official. Well, last week we had our 3 year anniversary together. Time flies.
Since my last post, I got accepted into the biotechnology program, for which I am forever grateful for. The program is very, very difficult and was the most mentally and emotionally taxing thing I had ever done up until that point. Being in class from 8:30am-4:30pm everyday with only 1 hour lunch break was rough. 6 to 7 classes a term was rough. Having a never ending midterm season was rough. But it was worth it. I made some amazing friends who I will hopefully have for the rest of my life. I gained some amazing lab skills and had some amazing opportunities. And best of all, I can actually see an end goal for my career. I don't know exactly what it will look like yet, but I know I want to be working on research in some sort of management position. Maybe not for forever, but for some time at least.
First year at BCIT was rough. Hard adjustments, lots of work and figuring out our class dynamic. But I came out of it stronger, knowing more about what I could do and how to succeed. Sean and I went through a rough patch in November of our first year together. His best friend from high school, whom he had feelings for for most of the time in school with her, was breaking up with her boyfriend of 5 years and basically went to Sean and said "I said no in the past but if you asked me now, I would say yes." And he doesn't know what to do so all he can do is come and tell me about it. And of course this happens on my birthday. So what do I do for my 18th birthday? Cry all night long. Yea, it was fun. It takes him a week to decide to choose us. Pretty rough but in the end it makes us stronger, especially since she comes around again in a year (but I'll get to that). After that Christmas, things change again. We were happy and then suddenly we stopped texting. Because of my schedule, we could only see each other once a week on Fridays. Everything was fine when we were together but during the week, everything felt empty and wrong. It took us until April to finally talk about it - I even wrote a letter about being upset that he's never read. But we finally talk about it and we figure out that we need to talk more and so we start calling each other. From then on, we try to discord each night and it has done us well.
After my first year at BCIT, I land my first co-op job in the Hancock Lab. I didn't think I was going to get a job at that point but I was so glad that I did. We did some really cool stuff with pseudomonas where we screened mutants for biofilm defects and tested biofilm growth in anaerobic conditions. I got to present my first poster at CBR Research Day. The lab pre-covid is amazing. Susan bought us beer and pizza and sushi every Friday and over the course of the summer, the drinking of the beer would start earlier and earlier (5pm at the beginning, all the way up till 2pm by the end of the summer). I had a great time in the lab and learned a lot.
My second year at BCIT was rough too. The workload got even worse that first term and I'm pretty sure we all hit our lowest lows. Just surviving became the name of the game and we did it well. Despite that, we had time to go play volleyball and support Josie's badminton tournament and fool around playing ultimate and snowball fights. We had some amazing memories and we didn't realize how fast time would fly until it was almost over.
And then Covid-19 hit. Assumed to be transferred from some sort of animal to humans in a Chinese wet market, racism and violence against Asians skyrocketed, just as the world shut down. Our last month at BCIT was canceled right before our eyes and we never got to celebrate finishing and surviving. Instead, classes went to online lectures and exams went to online formats and we stopped being able to see friends or go out or do anything really. Restaurants and attractions were shut down, maximum capacities and masks enforced and uncertainty everywhere. Talks for vaccines were hopeful, but I was skeptical about anything being ready until 2021. And I was right. As of today, all of us in the family have 1 dose of Moderna, although Mom is to get her second dose next week. Things are slowly opening up (provincial travel bans were lifted and movie theaters opened 2 weeks ago!) They're talking about what a post-covid world will look like, and I think everyone is grateful. In some ways, we lost a year and a half of our lives to this virus.
After finishing my time at BCIT, I was hoping to do a 4 month co-op placement abroad. Nothing of my applications turned out, but given covid, all travel ended up being restricted anyways. Not only that, there were no co-op jobs as every company in the world faced very uncertain economic and social times. I ended up taking April and May off and worked June and July at Collingwood again. Camp was different (lots of pool noodles and yoga mats) but in some ways, very much the same. I was grateful.
Despite the continued uncertainty of the next school year in a pandemic world, I was lucky to have the connections with the Hancock lab to allow me to do a full 8 month Honours Thesis with them. I took 4 classes per term on top of that and took them in the bioinformatics room on my laptop so I could be in the lab for the rest of the day. And boy, was I always there. 9am starts to 7pm finishes were not uncommon. Plus the 1.5 hour commute each way. Things were not easy. I thought BCIT was hard. 4th year at UBC trumps BCIT, easy. I was always stressed and strung out, I was constantly having to miss classes to do experiments (thank God for recordings) and at times, basic things refused to work (bacterial plating will be the death of me yet). The mandatory classes were all crappy and each have their own story that I may have to tell another time, but needless to say, I was not having a good time. I'm glad I made the effort to do an 8 month project, and in a way Covid made it both easier and harder. Easier because everything was recorded so my schedule was flexible (although I did my best to try to attend most lectures synchronously). Harder because I was in the lab more than I should have been and it meant some of my school work was compromised (thank God for Nabeel inviting me to his CHBE group). I survived, but I don't think I would have been able to go on much longer. Thank God for co-op and 8 months away from school.
In January, we were all on the hunt for co-op positions. Amazingly, I was super popular, scoring 6 different interviews of the 12 different applications I put in. I never got a Zymeworks invite though, which made me a little disappointed. But I think it worked out for the better because it meant I was more open when I was calling with Michelle. Meeting with Ting and Julien, we hit it off right away. I never get nervous for interviews and because of that, I feel like I'm really good at covering and clicking with interviewers. I asked lots of good questions and we were all laughing during that 1 hour interview. I got a call 2 days later from Michelle telling me I had gotten the job and I was so excited. It was the perfect fit. The chance to do more cell culture. The chance to do some research. The chance to maybe be hired on after grad. Working now, I feel really grateful for the opportunity. The company is a little odd, but our little CPD bubble is great to be a part of and I hope that one day we can make a difference.
I bought my first car on my first day of work - a 2012 Mazda 3 Hacthback with only 105k km. He's black and I've run him into the curb a few times, but he still looks super shiny (despite getting shat on within the first hour of being home) and I love him very much. The freedom of having my own car is amazing.
So why am I posting now? Well, I've been having some doubts and I needed a place to write it all out and I remembered this Tumblr. Reading back my old posts is a little sad because I started this Tumblr because I was sad. Things have been so much better in the last few years so I haven't been around. And make no mistake, I am so much happier than I was back then. But I've always learned while writing, so here I am to learn about myself and my feelings.
TBC
0 notes
Text
Aalto-Helsinki 2020 is here, come meet us!
The time has come for Aalto-Helsinki 2020 to finally take over the blog! Despite the COVID-19 situation, we have been busy with planning our project since February. We have brainstormed, read countless scientific articles, contacted experts and companies and had a lot of online meetings.
Now let us introduce ourselves:

Tytti
I did my Bachelor's degree in Biotechnology and Chemical Engineering and decided to continue with Biotechnology to the Master’s studies. What fascinates me the most about biotechnology is that we can study and engineer the tiniest components of life and at the same time do industrial scale production with them. I applied to iGEM because I’m very interested in entrepreneurship and this project mimics all the different elements that are needed to develop a business. Apart from science, my favourite things in life are playing the guitar, spending time in nature and enjoying tea.
An adventurous trip you have been on? My most recent adventurous trip was to Norway because I had always wanted to visit the Arctic Ocean. There was this fishing boat where some Finnish guy had designed a sauna among other unusual things. After warming up in that sauna, I jumped from the deck to the ice-cold water. It was cool to actually experience the Arctic Ocean fully!
Your most extraordinary skill? Making perfectly thin “muurikka” crepes
What is your favourite board game? Pandemic, rising tide version.

Carla
I am currently studying in the Ecology and Evolutionary Biology MSc program at the University of Helsinki. I completed my Bachelor’s degree in Genetics at Universitat Autònoma de Barcelona (UAB), in Barcelona, Spain. I have always been interested in Genetics and Evolution of organisms. I believe that a lot has still to be learnt about evolution mechanisms to make insights that can be applied to healthcare and medicine. In my free time I like to exercise, I am a bouldering enthusiast and I also try to go running as much as I can. I love watching movies as well. I applied to iGEM because it seemed the perfect opportunity to work with a multidisciplinary team and get to know people from different science fields. I consider myself an extroverted and curious person, so this could be the perfect opportunity to make new friends and learn from them. I thought it would also be the perfect opportunity to start a project on a topic of interest for all of us from scratch.
What is your favourite movie of all time? I don’t have a unique favorite movie, but my list of must watch movies is the following: Inception, Shutter Island, Green Book, The Good Will Hunting, The Shawshank Redemption, Jungle, Into the Wild, Mr. Nobody, Untouchable, Captain Fantastic, Interstellar, Donnie Darko, V for Vendetta and Dead Poets Society.
What kind of music do you listen to? I don’t only listen to one type of music, I like all types. Some international artists that I like are: Jonathan Wilson, John Frusciante, Steven Wilson, Eddie Vedder, Chet Faker, Gus Dapperton, Isaac Gracie, Billie Eilish, The Kooks, and Men I Trust among others. When talking about Catalan and Spanish music I usually listen to rumba, and rock, and also a bit of pop.
Your dream travel destination? My dream travel destinations would be: Iceland, Canada, Japan and Australia!

Amanda
I am studying at the Bachelor’s programme in Science in the chemistry study track at the University of Helsinki. In this year’s iGEM team I will work in the wet-lab but I am also involved with the funding, human practices and business. I applied to Aalto-Helsinki team to get the opportunity to take theory to practice and to learn a lot. In my free time I like cooking, baking, crocheting, knitting and gardening. I recently bought a greenhouse, where I will grow lots this summer!
What’s the most interesting or adventurous trip you have been on? Hard choice, but I would have to say the trip to Kenya with my family. So different and we saw a lot of animals and the whole Big Five!
What is your favourite board game now? And as a child? I think my favorite board game now is Dixit, but as a child I think it was Kimble.
Coffee or tea? Definitely tea, preferably green or white.

Artur
I study bioinformation technology at Aalto University. My minor is computer science. My role in our iGEM team is dry-lab and during the project I will be focusing on modelling as well as wiki development. I applied to iGEM because I wanted to be a part of a student driven team project and international competition. In my free time I enjoy programming, playing video games and going to the gym.
Your best tip to survive the quarantine? Outdoor activity, that does not require other people e.g. skateboarding and riding bicycle. Also playing video games with friends and watching tv series makes it easy.
What is your favourite movie of all time? Definitely Interstellar. After watching this for the third time I still get chicken skin. This almost three hour movie is a perfect combination of action, sci-fi and drama. Hans Zimmer did an astonishing job on music in this movie. I still keep one of the movie's soundtrack as my wake-up alarm. It's so good!
Who is your favourite scientist and why? Theoretical physicist Michio Kaku. I remember watching his show on Discovery Channel about black holes and teleportation. He got me interested in science and that's why he is my favourite scientist.

Daria
I am a second year master's student in Genetics and Molecular Biosciences, with focus on Molecular and Analytical Biosciences at the University of Helsinki. I have done my bachelor's in Biotechnology at the University of Silesia. I have experience in both environmental and biomedical research.
What was your favourite game to play as a child? Scavenger hunt. We lived right next to the forest, it felt good to be there for hours unsupervised.
What’s the grossest food you ever had to eat to be polite? Bananas. My culinary nemesis. Being grossed out by them is one of my earliest childhood memories.
What was the worst haircut you ever had? Bob at the age of 13. Thanks, mum.

Emilia
I have a Bachelor’s degree in Biotechnology from Tampere University and I’m now continuing my MSc (Tech) studies at Aalto University School of Chemical Engineering, major in Biotechnology. In my free time, I work out at the gym and love to cook. Also, I enjoy being outside in nature and spending time with my family and friends. I applied to iGEM because of my passion for life sciences and interest in challenging myself. I wanted to be a part of a powerful team of young professionals who share the same goal to achieve something meaningful. iGEM is a unique opportunity to work in an interdisciplinary team and carry out a research project from start to finish. In this iGEM project, my main responsibility is working in the wet-lab and obtaining funding. I will also contribute to human outreach activities and updating the team’s social media. Furthermore, I hope to learn more about bioinformatics and modeling. I have some experience in biomedical research and I’m looking forward to expanding my knowledge in the field of synthetic biology during the iGEM project. In the future, I wish to work with innovations which would somehow improve the quality of life.
What is your favorite time of the day and why? I love quiet mornings. I want to enjoy my morning coffee in peace and start my day without rushing. On the other hand, I also love spectacular sunsets, especially during the summer in the Finnish archipelago.
Your dream travel destination? Iceland. I would love to get to experience its unique nature, geysers and hot springs.
Coffee or tea? Coffee, of course.

Gustav
I am currently finishing my first year of masters studies in biosystems and biomaterials engineering. I applied for iGEM since I have always been interested in cell mechanisms and the intriguing and complex principles behind these cellular-systems. In the Aalto-Helsinki team, I am primarily doing wet-lab and budgeting, which means that I’m currently mostly researching data and cell-mechanisms related to our project. I often find one hobby at a time for which I am very passionate until I get tired of it and eventually switch. Previous hobbies have included been sewing, chess and origami, but for the moment I most do horticulture.
What is your dream travel destination? I would love to see the Socotra Island and the strange species living there.
What makes you happy? A cup of good quality tea in the evening.
Cats or dogs? Both??

Julia
I study Molecular Bioscience at the University of Helsinki. My part in iGEM is mainly in the wet lab, but I also take part in social media and human practises. Outside of iGEM I enjoy reading and listening to music, as well as playing video games and D&D. I applied to iGEM because I wanted to do something different and concrete with my knowledge and skills. Besides molecular biology for the last year I’ve been studying Chinese and hope to be fluent in it one day.
Most interesting trip you have been on? A couple of years ago my friends and I went on a trip to Chongqing, China. The trip was very pleasant despite none of the locals knowing English, the hole in the outer wall of the room or there being no warm water for the first couple of days. One thing led to another and we somehow ended up as models for the hostel’s website, which meant a full-blown photoshoot with free drinks and food. Afterwards the hostel owner even treated us to delicious hot pot!
Also, the baby pandas were cute.
What is your favourite book of all time? The Invisible Library by Genevieve Cogman. The heroine of the story is refreshingly sensible, the plot innovative and the book builds an interesting world for the sequels. Would recommend to anyone who likes fantasy, paranormal, or is interested in librarians using spycraft to steal books from alternative worlds.
Coffee or tea? Most definitely tea. Jasmine tea is especially close to my heart.

Maria
I am a third-year physics student from the University of Helsinki, majoring in theoretical physics and minoring in computer science. My main motivation for applying to iGEM was to apply my science skills to a real-world problem. I’m also eager to learn how research projects work. In our project I will focus on modelling, human practices and social media. Besides science, I love languages, especially French. In my free time I enjoy ballet, board games, walks and reading.
What fictional world would you love to visit? With all the chaos in the world currently, I’d love nothing more than to escape to Moominvalley. Moominmamma’s pancakes and a worry-free life sound really appealing. I’d also like to visit Hogwarts, I have been waiting for my letter since I was 10.
What skill would you love to master? I’d love to master flying, imagine all the freedom it would bring! However, all my attempts so far have failed. I wonder why that is.
What is your favourite board game? Probably Battlestar Galactica or Dale of Merchants.

Nata
I study Biosystems and Biomaterials Engineering in the program of Life Science Technologies at Aalto University. I completed my Bachelor's degree in Biotechnology and Chemical Technology. In this journey of iGEM, I'm responsible for Human Practice activities and taking part in the wet-lab work as well. In my free time, when I'm not passionately studying life sciences or being fascinated by nature, I keep myself active with group gymnastics, friends and family. I applied to iGEM to be part of the fascinating project in the field of synthetic biology. I'm sure that we will learn precious skills of planning, team working and presenting as well as meet many wonderful people! Currently beside the iGEM, I'm working with seedlings of tomatoes and herbs that I will relocate in my glass house when it gets warmer.
Describe your dream job: In my dream job, I would be able to utilize the field of life sciences to help people and the environment at the same time.
Your best tip to survive the quarantine? Your favourite movie of all time?
My tip to survive this quarantine would be to spend time in nature and watch as many movies as possible. One of the movies should definitely be my all time favourite: The Intouchables.
0 notes
Text
Big Data in Bioinformatics

Big data in action — A populated server room in a business data center. Adapted from a photo by Manuel Gessinger for Pexels.
When a biologist sets out to do research on something like the function of certain genes, they typically focus first on their main question: which genes affect a phenotype? They might then ask themselves what tools they’ll need to conduct experiments. Geneticists might use DNA sequencers, cell cultures, or assays. These techniques might be perfect for working on a single strain of bacteria or a single gene, but research today involves much larger tasks. With the rise of the information age, researchers have accumulated such an excess of research data that it’s no longer practical to manually analyze or sort through it.
This excess is widely known as big data. Science editor Vivien Marx of Nature magazine mentions that big data was once only relevant in astronomy and particle collider physics. However, the rapid advancement of technology in areas like genome sequencing has brought an explosion in data on the scale of petabytes. For scale, if laptops have 500 gigabytes of storage on average, a single petabyte can store 2000 times that volume. With such an exponential growth, how do biology researchers work with so much data?
How to train your algorithm
Before we can understand large-scale data on the order of petabytes, we must first look at the fundamentals of data analysis on a smaller scale. Algorithms―ways of managing and sorting information with modern computers―are a key factor in modern data analysis. The field that concerns algorithms is known as machine learning.
Traditionally, biologists perform “wet” lab work done by hand. Penders' genomics team studies nutrigenomics, which concerns the relationship between diet and genetics. One of the wet techniques they use is manually analyzing proteins with gels. Plotting genetic data after genome editing is another kind of wet lab work, as proteomics veteran Udeshi explains. But each case has something in common: they stop working after a certain point. Individually observing dots on a gel to identify proteins is only efficient until you have thousands of tests to perform, and plotting points only works as long as Excel doesn’t crash from data overload.

Gel electrophoresis — Common “wet” technique used to manually separate nucleic acids and proteins by mass. Taken from an IBBL article.
Despite how commonplace wet lab techniques are, Penders stresses that “dry” bioinformatics techniques can’t be left out of the picture in genomics. Computer algorithms are indispensable no matter which field a researcher studies, whether it be in Penders' nutrigenomics research or in Udeshi's proteomics work. Algorithms are unique since they facilitate sorting through large data sets and minimize wasted data. It can be easy for researchers to pick out the most relevant data and discard the rest, but algorithms are able to find associations invisible to the unaided eye.
When it comes to algorithms, statistical techniques are an essential part of the equation. Conventional methods of statistical analysis are best suited for data sets with less variables and are parametric, meaning they rely on bell-shaped, normal distributions. On the other hand, some statistical techniques are nonparametric. This special characteristic makes studying larger, multi-variable data sets easier, and lets researchers highlight relationships in noisy, ambiguous data sets.
For instance, Gonzalez-Recio's animal scientist team uses a nonparametric technique to study genomics in cows. This algorithmic technique, known as "gradient boosting" or “random boosting”, was devised by the researchers to better analyze subtle genetic differences called SNPs, or single nucleotide polymorphisms.
Of course, most algorithms require a base to learn from. In Gonzales-Recio's study, the boosting algorithm was fed a genomic test data set, only eventually gaining predictive ability after many iterations of machine learning.

Gradient boosting — Relies on gradual, sequential training with new models. The error circles represent discarded data that doesn't fit the model. By Bradley Boehmke from the University of Cincinnati.
Typically, chemists and biologists perform nuclear magnetic resonance or NMR on proteins in order to understand their structure, but this can be time-consuming and impractical on a larger scale. Another machine learning study by Kumar et al. involves feeding a chemical database into an algorithm to teach it how to classify proteins. The researchers' new algorithm provides an alternative to NMR by allowing low-level prediction without the need for NMR data. NMR still continues to be a mainstay lab technique for the foreseeable future, but this application of machine learning shows promise for the future of biochemistry and proteomics.
It’s easy to see now how computational algorithms and machine learning can reveal new perspectives in bioinformatics and genomics research. But only so much information can be analyzed by individual labs and computers. What happens when biologists need to deal with even larger sets of data?
Head in the clouds
Enter the era of cloud computing. The term cloud computing refers to rentable, customizable, online solutions for storage and processing power. The construct called The Cloud in this scenario is a network of decentralized, global data centers that allocate resources to users as needed. If this is difficult to picture, consider cloud services as virtual, remote-controlled, miniature computers. Although the field of cloud computing has only seen its advent in the past 2 decades, cloud computing is playing an increasingly crucial role in bioinformatics.
Cloud computing is magnitudes more powerful and efficient in comparison to local storage and analysis at individual labs or systems. With their expertise in computational biology, Langmead and Nellore emphasize the versatility of cloud computing in particular. Researchers can easily rent out and unload industry-grade resources at a relatively low cost, and tailored cloud services are available for every specific need. Companies such as Google and Amazon provide cloud solutions adapted for genomics and the life sciences as well.
Google Cloud — Cloud services can be used to deploy virtual, customizable computing units. Screenshot of a cloud control panel where users can customize their units. By Maks Osowski.
To illustrate, a genomics researcher might deploy a platform as a service (PaaS) instance to run RNA analysis software on. Not only would they be able to run their own set of tools on their own digital workspace, but they would also be able to reproduce and access their data anywhere. This confers special advantages for teams. Team members working on a genomics research project would be able to access the same virtualized workspace remotely, and manipulate data in real time.
Aside from on-demand cloud services, a variety of large databases exist online for genomic and genetic information. The National Center for Biotechnology Information, or NCBI, hosts a database called dbGaP with data gathered from research on phenotypes and genotypes, while gnomAD thoroughly documents genetic sequences along with statistical information and allelic frequencies. These databases may not represent big data on their own, but they serve as an important fraction in the big data equation. These databases contribute to the extensive, modern library of research data and serve as useful tools for biologists.

BLAST — A comprehensive database for genetic and protein sequences run by the NCBI. Screenshot of front page, featuring links to the search engines.
Resurfacing from a deep dive
Ultimately, many biologists who specialize in genomics, proteomics, and adjacent fields find themselves in a liminal space between the duality of “old” biology and “new” biology. Some may meet the idea of big data and bioinformatics with resistance. Others may welcome the change with open arms. But one thing is for sure: as researchers develop new technologies in bioinformatics and data science, researchers must adapt to working with increasingly larger data sets. The prevalence of big data forces older and newer biologists alike to realize the often-neglected importance of computer science and statistics.
It is undisputable that the new era of big data has brought much uncertainty, but in turn the era has also given us the rise of modern solutions. The advent of machine learning, algorithms, and cloud computing to automate processes that we would previously do manually has shifted the starting point of research from being inference-based to data-driven. Researchers are now able to compute results they would have never been able to a decade ago with their technological ceiling.
Whether or not these new technologies seem foreign to you, it's important to recognize that they aren't meant to be feared. As we make new discoveries in the world of science, we invent new approaches to how we research and test our ideas. Change breeds uncertainty and fear, but adaptation is an essential catalyst for future innovation and discovery. And if those discoveries end up changing us and our world in turn, we'd certainly be wise to welcome them with open arms. ■
0 notes
Text
300+ TOP Deep Learning Interview Questions and Answers
Deep Learning Interview Questions for freshers experienced :-
1. What is Deep Learning? Deep learning is one part of a broader group of machine learning techniques based on learning data analytics designs, as exposed through task-specific algorithms. Deep Learning can be supervised us a semi-supervised or unsupervised. 2. Which data visualization libraries do you use and why they are useful? It is valuable to determine your views value on the data value properly visualization and your individual preferences when one comes to tools. Popular methods add R’s ggplot, Python’s seaborn including matplotlib value, and media such as Plot.ly and Tableau. 3. Where do you regularly source data-sets? This type of questions remains any real tie-breakers. If someone exists going into an interview, he/she need to remember this drill of any related question. That completely explains your interest in Machine Learning. 4. What is the cost function? A cost function is a strength of the efficiency of the neural network data-set value with respect to given sample value and expected output data-set. It is a single value of data-set-function, non-vector as it gives the appearance of the neural network as a whole. MSE=1nΣi=0n(Y^i–Yi)^2 5. What are the benefits of mini-batch gradient descent? This is more efficient of compared tools to stochastic gradient reduction. The generalization data value by determining the flat minima. The Mini-batches provides help to approximate the gradient of this entire data-set advantage which helps us to neglect local minima. 6. What is mean by gradient descent? Gradient descent defined as an essential optimization algorithm value point, which is managed to get the value of parameters that reduces the cost function. It is an iterative algorithm data value function which is moves towards the direction of steepest data value function relationship as described by the form of the gradient. Θ: =Θ–αd∂ΘJ(Θ) 7. What is meant by a backpropagation? It ‘s Forward to the propagation of data-set value function in order to display the output data value function. Then using objective value also output value error derivative package is computed including respect to output activation. Then we after propagate to computing derivative of the error with regard to output activation value function and the previous and continue data value function this for all the hidden layers. Using previously calculated the data-set value and its derivatives the for output including any hidden stories we estimate error derivatives including respect to weights. 8. What is means by convex hull? The convex hull is represents to the outer boundaries of the two-level group of the data point. Once is the convex hull has to been created the data-set value, we get maximum data-set value level of margin hyperplane (MMH), which attempts to create data set value the greatest departure between two groups data set value, as a vertical bisector between two convex hulls data set value. 9. Do you have experience including Spark about big data tools for machine learning? The Spark and big data mean most favorite demand now, able to the handle high-level data-sets value and including speed. Be true if you don’t should experience including those tools needed, but more take a look into assignment descriptions also understand methods pop. 10. How will do handle the missing data? One can find out the missing data and then a data-set value either drop thorugh those rows value or columns value or decide value to restore them with another value. In python library using towards the Pandas, there are two thinging useful functions helpful, IsNull() and drop() the value function.

Deep Learning Interview Questions 11. What is means by auto-encoder? An Auto-encoder does an autonomous Machine learning algorithm data that uses backpropagation system, where that target large values are data-set to be similar to the inputs provided data-set value. Internally, it converts a deep layer that describes a code used to represent specific input. 12. Explain about from Machine Learning in industry. Robots are replacing individuals in various areas. It is because robots are added so that all can perform this task based on the data-set value function they find from sensors. They see from this data also behaves intelligently. 13. What are the difference Algorithm techniques in Machine Learning? Reinforcement Learning Supervised Learning Unsupervised Learning Semi-supervised Learning Transduction Learning to Learn 14. Difference between supervised and unsupervised machine learning? Supervised learning is a method anywhere that requires instruction defined data While Unsupervised learning it doesn’t need data labeling. 15. What is the advantage of Naive Bayes? The classifier preference converge active than discriminative types It cannot learn that exchanges between characteristics 16. What are the function using Supervised Learning? Classifications Speech recognition Regression Predict time series Annotate strings 17. What are the functions using Unsupervised Learning? To Find that the data of the cluster of the data To Find the low-dimensional representations value of the data To Find determine interesting with directions in data To Find the Magnetic coordinates including correlations To Find novel observations 18. How do you understanding Machine Learning Concepts? Machine learning is the use of artificial intelligence that provides operations that ability to automatically detect further improve from occurrence without doing explicitly entered. Machine learning centers on the evolution of network programs that can access data and utilize it to learn for themselves. 19. What are the roles of activation function? The activation function means related to data enter non-linearity within the neural network helping it to learn more system function. Without which that neural network data value would be simply able to get a linear function which is a direct organization of its input data. 20. Definition of Boltzmann Machine? Boltzmann Machine is used to optimize the resolution of a problem. The work of the Boltzmann machine is essential to optimize data-set value that weights and the quantity for data Value. It uses a recurrent structure data value. If we apply affected annealing on discrete Hopfield network, when it would display Boltzmann Machine. Get Deep Learning 100% Practical Training 21. What is Overfitting in Machine Learning? Overfitting in Machine Learning is described as during a statistical data model represents random value error or noise preferably of any underlying relationship or when a pattern is extremely complex. 22. How can you avoid overfitting? Lots of data Cross-validation 23. What are the conditions when Overfitting happens? One of the important design and chance of overfitting is because the models used as training that model is the same as that criterion used to assess the efficacy of a model. 24. What are the advantages of decision trees? The Decision trees are easy to interpret Nonparametric There are comparatively few parameters to tune 25. What are the three stages to build the hypotheses or model in machine learning? Model building Model testing Applying the model 26. What are parametric models and Non-Parametric models? Parametric models remain these with a limited number from parameters also to predict new data, you only need to understand that parameters from the model. Non Parametric designs are those with an unlimited number from parameters, allowing to and flexibility and to predict new data, you want to understand the parameters of this model also the state from the data that has been observed. 27. What are some different cases uses of machine learning algorithms can be used? Fraud Detection Face detection Natural language processing Market Segmentation Text Categorization Bioinformatics 28. What are the popular algorithms for Machine Learning? Decision Trees Probabilistic networks Nearest Neighbor Support vector machines Neural Networks 29. Define univariate multivariate and bivariate analysis? if an analysis involves only one variable it is called as a univariate analysis for eg: Pie chart, Histogram etc. If a analysis involves 2 variables it is called as bivariate analysis for example to see how age vs population is varying we can plot a scatter plot. A multivariate analysis involves more than two variables, for example in regression analysis we see the effect of variables on the response variable 30. How does missing value imputation lead to selection bias? Case treatment- Deleting the entire row for one missing value in a specific column, Implutaion by mean: distribution might get biased for instance std dev, regression, correlation. 31. What is bootstrap sampling? create resampled data from empirical data known as bootstrap replicates. 32. What is permutation sampling? Also known as randomization tests, the process of testing a statistic based on reshuffling the data labels to see the difference between two samples. 33. What is total sum of squares? summation of squares of difference of individual points from the population mean. 34. What is sum of squares within? summation of squares of difference of individual points from the group mean. 35. What is sum of squares between? summation of squares of difference of individual group means from the population mean for each data point. 36. What is p value? p value is the worst case probability of a statistic under the assumption of null hypothesis being true. 37. What is R^2 value? It’s measures the goodness of fit for a linear regression model. 38. What does it mean to have a high R^2 value? the statistic measures variance percentage in dependent variable that can be explained by the independent variables together. 40. What are residuals in a regression model? Residuals in a regression model is the difference between the actual observation and its distance from the predicted value from a regression model. 41. What are fitted values, calculate fitted value for Y=7X+8, when X =5? Response of the model when predictors values are used in the model, Ans=42. 42. What pattern should residual vs fitted plots show in a regression analysis? No pattern, if the plot shows a pattern regression coefficients cannot be trusted. 43. What is overfitting and underfitting? overfitting occurs when a model is excessively complex and cannot generalize well, a overfitted model has a poor predictive performance. Underfitting of a model occurs when the model is not able to capture any trends from the data. 44. Define precision and recall? Recall = True Positives/(True Positives + False Negatives), Precision = True Positives/(True Positives + False Positive). 45. What is type 1 and type 2 errors? False positives are termed as Type 1 error, False negative are termed as Type 2 error. 46. What is ensemble learning? The art of combining multiple learning algorithms and achieve a model with a higher predictive power, for example bagging, boosting. 47. What is the difference between supervised and unsupervised machine learning algorithms? In supervised learning we use the dataset which is labelled and try and learn from that data, unsupervised modeling involves data which is not labelled. 48. What is named entity recognition? It is identifying, understanding textual data to answer certain question like “who, when,where,What etc.” 49. What is tf-idf? It is the measure if a weight of a term in text data used majorly in text mining. It signifies how important a word is to a document. tf -> term frequency – (Count of text appearing in the data) idf -> inverse document frequency tfidf -> tf * idf 50. What is the difference between regression and deep neural networks, is regression better than neural networks? In some applications neural networks would fit better than regression it usually happens when there are non linearity involved, on the contrary a linear regression model would have less parameters to estimate than a neural network for the same set of input variables. thus for optimization neural network would need a more data in order to get better generalization and nonlinear association. 51. How are node values calculated in a feed forward neural network? The weights are multiplied with node/input values and are summed up to generate the next successive node 52. Name two activation functions used in deep neural networks? Sigmoid, softmax, relu, leaky relu, tanh. 53. What is the use of activation functions in neural networks? Activation functions are used to explain the non linearity present in the data. 54. How are the weights calculated which determine interactions in neural networks? The training model sets weights to optimize predictive accuracy. 55. which layer in a deep learning model would capture a more complex or higher order interaction? The last layer. 56. What is gradient descent? It comprises of minimizing a loss function to find the optimal weights for a neural network. 57. Imagine a loss function vs weights plot depicting a gradient descent. At What point of the curve would we achieve optimal weights? local minima. 58. How does slope of tangent to the curve of loss function vs weigts help us in getting optimal weights for a neural network Slope of a curve at any point will give us the direction component which would help us decide which direction we would want to go i.e What weights to consider to achieve a less magnitude for loss function. 59. What is learning rate in gradient descent? A value depicting how slowly we should move towards achieving optimal weights, weights are changedby the subtracting the value obtained from the product of learning rate and slope. 60. If in backward propagation you have gone through 9 iterations of calculating slopes and updated the weights simultaneously, how many times you must have done forward propagation? 9 61. How does ReLU activation function works? Define its value for -5 and +7 For all x>=0, the output is x, for all x Read the full article
0 notes
Text
Paper annotations #1
tmVar 2.0: integrating genomic variant information from literature with dbSNP and ClinVar for precision medicine
Intro
Each SNP record in dbSNP (Database for Short Genetic Variations) is assigned a stable and unique variant accession identifier (RSID), which is linked to aggregated information (associated gene, functional consequences and allele frequency).
NHGRI-EBI GWAS Catalog is a collection of genome-wide set of genetic variants in different individuals associated with a trait [1].
For genomic variant information in cancer, COSMIC contains expert-curated data of somatic mutations [2].
CIViC is an open-acess, open-source knowledgebase for expert-crowdsourced of clinical interpretation of variants in cancer [3].
DisGeNET is a recent platform integrating information on gene-variation-disease associations from several public data sources and the literature [4].
"The first version of tmVar is a high-performance software for external evaluations comparing formats in the PubMed article and re-writing them in HGVS formats (e.g. p.Pro12Ala). However, HGVS names can still be ambiguous: one can often be linked to multiple RSIDs (e.g. rs767209585 and rs773973301 are both associated with p.Pro12Ala). Indeed, on average, one protein mutation in HGVS name maps to more than ten RSIDs".
Why not use HGVS genomic nomeclature? HGVS isn't just the protein nomeclature, it considers the gene, genomic location and protein location.
"in this work we first extended tmVar to automatically normalize the variant mentions and map them to standard dbSNP RS numbers."
It includes variants not present in dbSNP that could be considered rares?
Using the human gold standard they compare tmVar 2.0 against SETH, another automated tool to text-mining mutations [5] and had nearly 90% in F-measures.
about F1
"Our analysis includes: (i) comparing the text-mined PMID-RSID pairs with annotated dbSNP data, (ii) analyzing variants curated in ClinVar and (iii) discovering novel connections between variants, gene and diseases"
"Our investigation revealed 161 178 missing RSID-PMID links in dbSNP and 41 889 RSIDs not found in ClinVar. Moreover, our results also include over 120 000 rare variants (MAF 0.01) in nearly 4000 genes across the genome which are presumed to be deleterious and are not frequently found in the general population."
MAF isn't enough to considered a variant patogenic, maybe more information had been considered
Materials and methods
"tmVar applies ML approach to tag mutation mentions in free txt, detecting terms that represent variants of multiple types (SNV, insertion, deletion, etc) and sequence context (genomic, transcript and protein) and returns its results in HGVS form".
"Before we performed normalization, we first built a comprehensive lexicon containing all possible mappings between variant mentions and RSIDs, harvested from three difference sources: dbSNP, Clinvar and PubMed".
Two main strategies were used to find corresponding RSID: pattern matching '[Gene/Protein] ([DNAMutation] with [RSID])' and a list of candidate RSIDs for search using lexicon. For disambiguation, they use global information in the entire article and/or variant-associated gene information, also using GNormPlus an end-to-end and open source system that handles both gene mention and identifier detection.
The frequency data used as population frequency come from 1000 Genomes Phase 3, Exac, NHLBI GO ESP and gnomAD.
Results
"The tmVar RS results (62452 RS numbers in 9782 genes) were categorized using dbSNP and ClinVar annontations along multiple facets, including functional consequences (syn, non-syn, etc) based on RefSeq mRNA annotations, minor allele frequency (MAF), and clinical significance in order to prioritize their biological significance and assess their clinical impact".
Discussion
According to the table 4, OSIRIS had better results than tmVar2. So, OSIRIS could be used with tmVar2.
"our results could be used by other computational methods in bioinformatics research such as connecting genotypes with phenotypes and/or modeling gene-disease-variant relations [DisGeNeT][6]"
2 notes
·
View notes
Text
Snapshot - San Francisco
I thought maybe I’d write a funny little poem about corgis and post it on Facebook. As is usually the case, I’ll try to embed something deep into humor...as is usually the case, I’ll probably fail. I think that the truly “deep” work I see usually just takes something real, and reflects it back.
Like this one blog post I reblogged recently...it was someone discussing how as a child, she felt that she was just pretending to like things that it was normal for a kid to like. This could be really deep and loaded with meaning, but the point is that it doesn’t have to be. It’s true. It just is.
So what is it I’m trying to convey?
I saw a lot of corgis today at the beach. Okay, sure, whatever. That’s really not something that I care that much about. They’re cute. If nothing else, I like that everyone seems to appreciate them.
What meant a lot more to me was walking through Embarcadero. Seeing tech central spread out around these little homeless populations. Lights changing. The look of the Bay Bridge over the water. I passed through Haight and spent yesterday there, and even though in a way it’s kind of shit I still really think that the area is San Francisco. It’s alive. It will go out in style. I’m thinking about that, and I’m thinking about me, and I don’t think I really am any of those things that are San Francisco. I guess that maybe someday I can come back as the tech component, but in my mind that never was San Francisco. That was something external, something that came from outside and slowly crept in. I don’t like or dislike that this is what much of the city became. I just don’t think of it as the essence.
And then there was a coffee shop where I met someone, by coincidence, whom I haven’t talked to in years. She’s moving to socal. She said she feels like we’re basically 30, even though we’re not. I asked if she’s thinking about marriage with someone first or a house first, and I said it like a joke even though it’s not.
That’s what’s really on my mind, and I know it doesn’t really have to mean anything. Tonight was my last night, but not really...I’m sure I’ll be back. I saw a bioinformatics company on the inside, and tons of familiar tech scenes I used to spend so much time in during my internship, and lots of little things that I think of as part of the city I know and love.
And that’s it. That’s all I care about for now. I have the experiences, even though sometimes I feel like it’s all just happening with or without me and I’m just this one introvert surrounding himself with people to see what it’s all about.
So, because I need a point of reference, why not just focus on corgis?
Where else is there to go?
2 notes
·
View notes