#protein structure
Explore tagged Tumblr posts
Note
Could you do the full lyrics to "we didn't start the fire"? Thanks in advance!
this feels fitting enough given the [everything in current events oh my god].
i used the original version of the song for this one, and i'm very glad i'm using AF3 since this is just over 2,000 characters!
letter sequence in this ask matching protein-coding amino acids:
HarryTrmanDrisDayRedChinahnnieRaySthPacificWalterWinchelleDiMaggieMcCarthyRichardNinStdeakertelevisinNrthKreaSthKreaMarilynMnreRsenergsHmSgarRayPanmnmrandTheKingandIandTheCatcherintheRyeEisenhwerVaccineEnglandsgtanewqeenMarcianLieraceSantayanagdyeWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefireNwedidntlightittwetriedtfightitsephStalinMalenkvNasserandPrkfievRckefellerCampanellaCmmnistlcRyChnanPernTscaniniDacrnDienienPhfallsRckArndtheClckEinsteinamesDeanrklynsgtawinningteamDavyCrckettPeterPanElvisPresleyDisneylandardtdapestAlaamaKrshchevPrincessGracePeytnPlaceTrleintheSeWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefireNwedidntlightittwetriedtfightitLittleRckPasternakMickeyMantleKeracSptnikChEnLairidgentheRiverKwaiLeannCharlesdeGalleCalifrniaaseallStarkweatherhmicidechildrenfthalidmideddyHllyenHrspacemnkeymafiaHlahpsCastrEdselisangSyngmanRheePaylaandKennedyChyCheckerPsychelgiansintheCngWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefireNwedidntlightittwetriedtfightitHemingwayEichmannStrangerinaStrangeLandDylanerlinayfPigsinvasinLawrencefAraiaritisheatlemanialeMisshnGlennListneatsPattersnPpePalMalclmritishpliticianseFKlwnawaywhatelsedIhavetsayWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefireNwedidntlightittwetriedtfightitirthcntrlHChiMinhRichardNinackagainMnshtWdstckWatergatepnkrckeginReaganPalestineterrrntheairlineAyatllahsinIranRssiansinAfghanistanWheelfFrtneSallyRideheavymetalsicideFreigndetshmelessvetsAIDScrackernieGetHypdermicsntheshreChinasndermartiallawRckandrllerclawarsIcanttakeitanymreWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefiretwhenwearegneItwillstillrnnandnandnandnandnandnandnandnWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefireNwedidntlightittwetriedtfightitWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefireNwedidntlightittwetriedtfightitWedidntstartthefireItwasalwaysrningsincethewrldseentrningWedidntstartthefireNwedidntlightittwetriedtfightit
protein guy analysis:
this turned out a lot better than i expected! there are a couple long loops that seem to be slightly weirdly positioned and the middle part is a bit of a weird shape that almost makes me feel as though the structure is missing a binding partner, but it would take an absolutely insane coincidence for this to be even a halfway decent fit for any evolved protein. still, if you look at it from far enough away, it almost looks like a generic enough globular protein. as always, i don’t actually trust or believe in any of this, but i guess it’s more interesting to look at than a mess of random coils?
predicted protein structure:

cartoon representation

surface representation
#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein asks#protein songs
70 notes
·
View notes
Text
raw input: penis filtered input: PENIS it's just a wibble, not much else to say i think?



stick | cartoon | surface

colored by pLDDT
110 notes
·
View notes
Link
OpenFold, an artificial intelligence (AI) research consortium, has announced the release of two new tools that aim to improve protein structure prediction: SoloSeq and OpenFold-Multimer. These open-source tools represent important advances in integrating protein language models with structure prediction software, as well as modeling protein complexes more accurately.
The SoloSeq model developed on Amazon Web Services (AWS) emerges as a groundbreaking protein LLM/structure prediction AI tool. It holds the distinction of being the inaugural fully open-source model in this category, offering critical training code. Now, organizations can leverage this code for fine-tuning or training new models with their proprietary data, marking a pivotal step towards collaborative advancements in the field.
The first tool, SoloSeq, combines OpenFold’s existing protein structure prediction capabilities with a new protein large language model (LLM). This integration eliminates the need for a separate pre-processing step to generate a multiple sequence alignment (MSA) that is required by OpenFold and other structure predictors like AlphaFold.
Continue Reading
21 notes
·
View notes
Text
Intrinsic Membrane Proteins

Patreon
#studyblr#notes#medblr#medical notes#med notes#cytology#cytology notes#biology#bio#human bio#microbiology#microbio#anatomy and physiology#cell biology#anatomy#physiology#proteins#biochemistry#biochem#cell biochemistry#cell structure#protein structure#cell proteins#insulin receptors#insulin#transmembrane proteins#intrinsic proteins#membrane proteins#organic chemistry#mcat
5 notes
·
View notes
Text
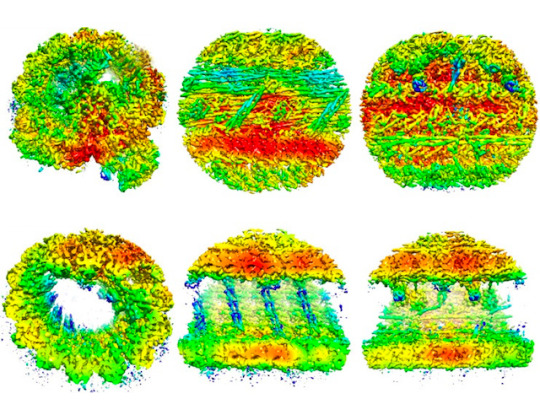
Building a Sperm
Combining protein structure predictions with cryoelectron tomography reveals proteins – novel and known – their structures, interactions and locations in intact mammalian sperm
Read the published research paper here
Image from work by Zhen Chen and colleagues
University of California San Francisco, San Francisco, CA, USA
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in Cell, October 2023
You can also follow BPoD on Instagram, Twitter and Facebook
5 notes
·
View notes
Text
this is it everyone. time to fully develop brain !!
fiveyearplanfckitweallfllydevelprainifstillinsanekillselfifnrmalstartpdcastwerernningtftimetfckitweall
letter sequence in this ask matching protein-coding amino acids:
fiveyearplanfckitweallfllydevelprainifstillinsanekillselfifnrmalstartpdcastwerernningtftimetfckitweall
protein guy analysis:
the confidence isn't great, but i do like how this one turned out. there are four nice helices, all arranged together in a way that looks like it could be a real domain. this is off topic, but sometimes i wonder how well the structures of protein interfaces that interact with other things are handled. for instance, if something was a transmembrane domain, or had a hydrophobic interaction with another subunit. i guess it would depend more than anything on how many similar structures there were, since i don't know how much the program looks at thermodynamics vs existing structures. i know at least that there are programs to predict transmembrane domains, i'm sure this is something i could research a little more instead of just making guesses. ultimately though, while there are some hydrophobic patches on the surface, they are weirdly dispersed in a way that makes me think this should be either a section of a full protein, or that this is actually just a mistake of language and computation that i have invested too much thought into.
predicted protein structure:

cartoon structure

surface, coloured by hydrophobicity (blue is most hydrophilic and yellow is most lipophilic, with white in the middle)

we're running out of time to fuck it we ball....
#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein memes#five year plan#happy new year!
207K notes
·
View notes
Note
the lyrics of family reunion by blink 182
that was,,, not what i expected
letter sequence in this ask matching protein-coding amino acids:
ShitpissfckcntccksckermtherfckertitsfarttrdandtwatShitpissfckcntccksckermtherfckertitsfarttrdandtwatShitpissfckcntccksckermtherfckertitsfarttrdandtwatShitpissfckcntccksckermtherfckertitsfarttrdandtwatIfckedyrmmAndIwannasckmydadAndmymmmythisthisthingn
protein guy analysis:
we really are just inventing new versions of amyloid fibrils over here. these cross beta structures (two parallel beta sheets, like what i'm seeing here - google it for real images and examples) feel so evil. i know its probably the mix of highly repetitive sequences and the bias towards structured proteins in the PDB, but i miss the spaghetti messes i used to get. i'll admit that i was too hard on those, and i'm currently taking a class that has talked more about intrinsically disordered proteins, which has given me a much better appreciation for them. oh well. an inevitable part of running this blog as i learn and take classes is that eventually i'll realize the flaws in my old posts. as always if i make a mistake or say something questionable, please correct me! no one is infallible and pretending otherwise is pointless. my one reassurance about this structure is that the confidence is low, so this probably would look messier for real.
predicted protein structure:

#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein asks#protein songs
152 notes
·
View notes
Note
the missile is very tired. he is eepy. the missile has had a very long day of splashing bandits and wants to take just a small sleep. he eeby and neebies to sleebie. mibsile sleepy and need bed by time. the missile is currently experiencing critical levels of being a sleehjy little guy and needs to go to beb. he is retired and needs to slep. just a little sleejing time as a treat. mibsilelelele neebs to slek for twired boyo. just a lil guy. mibsipaleebeelee needs his beaty sleep. look at him go! he yawn bib cause he skeegy. neebs to falafel asleep. ni ni time. goodnight, mr the missile.
raw input: ↑ filtered input: themissileisverytiredheiseepythemissilehashadaverylngdayfsplashinganditsandwantsttakestasmallsleepheeeyandneeiestsleeiemisilesleepyandneededytimethemissileiscrrentlyeperiencingcriticallevelsfeingasleehylittlegyandneedstgteheisretiredandneedstslepstalittlesleeingtimeasatreatmisileleleleneestslekfrtwiredystalilgymisipaleeeeleeneedshiseatysleeplkathimgheyawnicaseheskeegyneestfalafelasleepninitimegdnightmrthemissile theres a shit ton of alpha helices and a few beta sheets, it looks mostly orange, kinda stringy around the edges, kind of standard for a peptomination. yet, i somehow feel like it's on the good side of standard? although from a certain angle theres like. a hole. why is there a hole did the missile do that




cartoon | surface cartoon hole | surface hole
colored by pLDDT

13 notes
·
View notes
Link
Centuries after the first proteins were identified and characterized, their complexity proves a challenge for scientists all over the world. Their creation, evolution, and many variations are questions for which we have only just begun to uncover the answers. A new bioinformatics tool has now been developed that can provide researchers with a way to analyze data and map protein sequences and structures on a large scale. This has important implications for protein modeling and sequencing technologies.
Genomes provide the template on which all life is created and maintained. Despite the high accuracy of DNA replication mechanisms found in living organisms, certain mutations can still occur and be passed on. Such mutations can change the function and structure of the protein that the gene is supposed to encode. Nearly all Mendelian diseases are caused by variations in the proteome.
In lieu of the time-consuming and exhaustive process of determining protein structures and mutations in the lab, researchers have now turned to bioinformatics as an alternative method to quickly and cheaply generate highly accurate and precise protein structure models. Many protein modeling tools have been developed to predict mutations and their impact with as much accuracy and precision as possible. The majority of such tools use only protein sequencing information from both the protein as well as its variants in order to produce results. While such a method is advantageous in that it is applicable to nearly all genetic mutations, they don’t account for the structure of the mutated variants, resulting in a loss of valuable information that can be garnered from taking the proteins’ 3-dimensional structures into account. This is especially important as it allows for distinguishing between mere correlations in mutation occurrence and causative variations. Hence, the results obtained through such methods may not be as accurate or precise.
Continue Reading
25 notes
·
View notes
Text
finally, i've nailed it on the timing of a post!
letter sequence in this ask matching protein-coding amino acids:
daystandIstillthinkImin
protein guy analysis:
it looks like AF3 just loves helices. in this case at least i do trust the helix, and honestly i'm pretty blown away by the accuracy of this, and the prediction is giving me >90% accuracy!! smaller structures do usually turn out better, but even then this feels to good to be random, so i did a quick BLAST search. sure enough, i found a 91% match for most of the sequence from a bacteria enzyme (specifically aldose 1-epimerase from Neobacillus drentensis), and another match for the last part. between these two proteins, we have nearly complete coverage of the sequence, and even if neither of these have solved structures, there are a number of other similar results, so this doesn't look too far off from evolved sequences (albeit just for a small peptide fragment). this lines up nicely with the strengths of AlphaFold, since it can use existing information to make a fairly trustworthy and reasonable guess. even then, this could only be trusted so much without any experimental structural information.
predicted protein structure:

cartoon representation, coloured by pLDDT

cartoon with side chains, coloured by pLDDT

highest % identity BLAST result

another strong BLAST result

pLDDT colour code
16 days to 2015 and I still think I’m in 2012
#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein memes#BLAST#protein BLAST
162K notes
·
View notes
Text
AlphaFold Nobel Prize!
Hey everyone :) this isn't a structure, but there is some protein news that is pretty relevant to this blog that I felt I had to share. This article gives a nice overview of AI-predicted protein structures and what sorts of things they can do for research. It's not too long, and I recommend taking a look
If you've been seeing my posts for any amount of time, I've absolutely given you a flawed view of how useful AF can be. Experimentally determining protein structures is a demanding and difficult process (I've never done it, but I've learned the overview of how x ray crystallography works, and I can only imagine how much work it would take). AI-generated structures are not going to make structural biology obsolete, but they are massively helpful in making predictions that go on to guide further research.
While in many fields (especially creative areas like art and writing) AI has significant ethical concerns, I feel like this sort of use of AI in science is an overwhelmingly positive thing. The data used to train it is publicly available, and science works by building on the work done by those before us. Furthermore, while AI may not be great at generating new ideas or copying humans, it is very good at sorting large amounts of data and using it to make predictions. It's more akin to very complicated statistics than an attempt at the Turing test, and in this case it is a valuable tool to expand the ways we can do science!
#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein info
166 notes
·
View notes
Text
raw input: [the vaporeon copypasta]
filtered input: [the vaporeon copypasta but without the numbers, punctuation, spaces, and the letters b, x, j, z, o, & u. i'm not putting it here you can't make me]
a nice amount of secondary structure in the center, even including a few beta sheets! shame about the absolute spaghetti outside and through it. also 2 of the alpha helices look... crimped, in a word. a distressing peptomination for a distressing string of text. good thing neither are real like look how fuckign oragne it is. the surface view kinda looks like one of those things you get by pouring molten metal in an anthill and waiting for it to cool jesus fucking christ
(also. did you know ChimeraX has a Spin Movie feature? because i didn't)


cartoon | surface
colored by pLDDT

8 notes
·
View notes
Note
Let's try the legendary Brain Power chorus:
Let the bass kick
O-0000O00000 AAAAE-A-A-|-A-U- JO-
O0000O00000O AAE-O-A-A-U-U-A- E-eee-ee-eee AAAAE-A-E-I-E-A- JO-000-00-00-oo EEEEO-A- AAA-AAAA
O-o000o00000 AAAAE-A-A-l-A-U-JO-
O00000000000 AAE-O-A-A-U-U-A- E-eee-ee-eee AAAAE-A-E-|-E-A-
JO-o00-00-00-0o EEEEO-A-AAA-AAAA
O-o00O00000o AAAAE-A-A-|-A-U- JO-
O0000O00000o AAE-O-A-A-U-U-A- E-eee-ee-eee AAAAE-A-E-I-E-A-
JO-000-00-00-0o EEEEO-A-AAA-AAAA-
O---------
oh i am very interested to see how this turns out
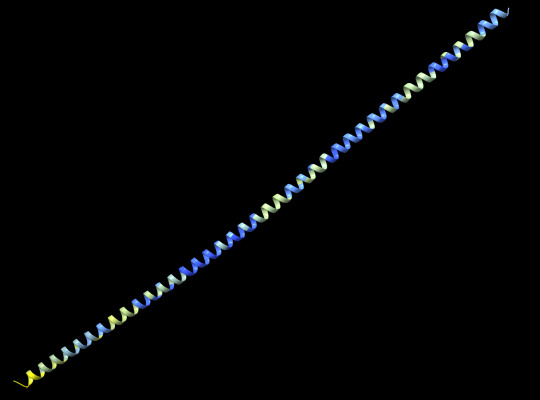
letter sequence in this ask matching protein-coding amino acids:
LettheasskickAAAAEAAAAAEAAAEeeeeeeeeAAAAEAEIEAEEEEAAAAAAAAAAAAEAAlAAAEAAAEeeeeeeeeAAAAEAEEAEEEEAAAAAAAAAAAAEAAAAAEAAAEeeeeeeeeAAAAEAEIEAEEEEAAAAAAAA
protein guy analysis:
well this is a long one. and by that i mean that the structure itself is just one long helix. for what it's worth, the confidence is pretty good. this makes enough sense, given that both alanine (A) and glutamate (E) tend to prefer to be in alpha helices. overall, this is just a silly little guy and i'm very happy with it
predicted protein structure:
#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein asks
167 notes
·
View notes
Text
my cells are more productive than i will ever be, and i commend the forces of nature for everything they have accomplished
letter sequence in this post matching protein-coding amino acids:
affirmatinsiamacmplerganismrtallyengineeredyncaringfrcesfnatreiamaprdctfillinsfyearsandtrillinsfdeathsiamildingamachinegreaterthanmyselfiamaletmakephnecallsandappintments
protein guy analysis:
there is a beta sheet and an alpha helix, but still a pretty standard amount of disordered loops around these structures. as much as i had complained about them before, i think its somewhat comforting to see some disordered loops along with the low confidence scores, since i don't really believe a lot of the structures that come out of these predictions. there are still some hydrophobic residues that are a little too exposed for my comfort, and like many of the peptonominations that this blog generates, i think this would be a challenge to try and express in cells. i don't think i've talked enough about aggregation, or how protein folding in vitro is different from in vivo, but i would be very interested to see how this interacts with a solvent or the mess of the cytosol
predicted protein structure:

cartoon representation

surface coloured by hydrophobicity
affirmations
i am a complex organism brutally engineered by uncaring forces of nature
i am a product of billions of years and trillions of deaths
i am building a machine greater than myself
i am able to make phone calls and appointments
66K notes
·
View notes
Note
Hi, im sorry if this violates no nsfw but i thought i might try coz its been an old meme; delete if you dont want to ans it, no pressure!
I've come to make an announcement: Shadow the Hedgehog's a bitch-ass motherfucker. He pissed on my fucking wife. That's right. He took his hedgehog fuckin' quilly dick out and he pissed on my FUCKING wife, and he said his dick was THIS BIG, and I said that's disgusting. So I'm making a callout post on my Twitter.com. Shadow the Hedgehog, you got a small dick. It's the size of this walnut except WAY smaller. And guess what? Here's what my dong looks like. That's right, baby. Tall points, no quills, no pillows, look at that, it looks like two balls and a bong. He fucked my wife, so guess what, I'm gonna fuck the earth. That's right, this is what you get! My SUPER LASER PISS! Except I'm not gonna piss on the earth. I'm gonna go higher. I'm pissing on the MOOOON! How do you like that, OBAMA? I PISSED ON THE MOON, YOU IDIOT! You have twenty-three hours before the piss DROPLETS hit the fucking earth, now get out of my fucking sight before I piss on you too!
tbh i'm not going to be super strict about the no nsfw thing, but i also don't entirely trust the internet and wanted to make sure no one would submit anything truly vile. this is more comedic than anything else though, so you're all good!
letter sequence in this ask matching protein-coding amino acids:
IvecmetmakeanannncementShadwtheHedgehgsaitchassmtherfckerHepissednmyfckingwifeThatsrightHetkhishedgehgfckinqillydicktandhepissednmyFCKINGwifeandhesaidhisdickwasTHISIGandIsaidthatsdisgstingSImmakingacalltpstnmyTwittercmShadwtheHedgehgygtasmalldickItsthesiefthiswalnteceptWAYsmallerAndgesswhatHereswhatmydnglkslikeThatsrightayTallpintsnqillsnpillwslkatthatitlksliketwallsandangHefckedmywifesgesswhatImgnnafcktheearthThatsrightthisiswhatygetMySPERLASERPISSEceptImntgnnapissntheearthImgnnaghigherImpissingntheMNHwdylikethatAMAIPISSEDNTHEMNYIDITYhavetwentythreehrsefrethepissDRPLETShitthefckingearthnwgettfmyfckingsightefreIpissnyt
protein guy analysis:
if you've been here long enough or have a general idea of what proteins are supposed to look like, i'm sure you could write this analysis from looking at the picture for about two seconds. all i have to say is this: why is it so spread out??? the answer, of course, is that all of the many polar and charged residues are way too hydrophilic to make anything real, and instead are just flopping around uselessly with all the water. proteins were never meant to have this much entropy, and this chaos is downright infuriating
predicted protein structure:

#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein asks#protein memes#shadow the hedgehog
349 notes
·
View notes
Text
Would it be possible to kill bacteria without damaging their protein structure?
Would it be possible to kill bacteria without damaging their protein structure? There are several ways to kill bacteria without damaging protein structure or function. For example, consider the following frequent ones: 1. First, heat inactivation: Although heat may be used to destroy bacteria, it can also denature proteins if it is applied at too high a temperature for too long. However, some…
View On WordPress
#Antibiotics#bacteria#Cchemical Fixation#gamma irradiation#heat inactivation#protein structure#UV Irradiation
0 notes