#or rather I spend 4600 words talking about a quarter of a scene as a springboard to talk about my job and how cool it is
Explore tagged Tumblr posts
Text
13/30 Science interlude!
(Previous) | (Index) | (Next)
⛬
We return to Prometheus, where I am taking a break to ramble about my job. A thing that I love. It will be a nice change. Also: weird blood!
I have been informed that some methods of accessing tumblr do not play well with long alt text rambles. To keep the flow between the main text and alt text separate, I’ll be copying the longest ones below the main text and citations. Captions that I think are going to be long enough to need this treatment will be marked with “Overflow Ramble [number]”, so they’ll be slightly easier to find. It’s not a perfect system, but Tumblr is not a perfect website.
And I am going to need the overflow space this time, because we’re getting into genetics!
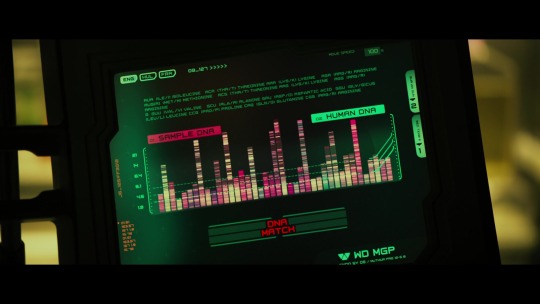
After electrocuting a decapitated alien head until it exploded into a shower of green gore, the creatures that claim to be scientists stuck a bit of the goop in some sort of very science-y DNA machine, leading to this:
“Let's have a look at its DNA. Isolate the strand. Okay. Compare it to the gene sample?” “[Overlay… Processing… Processing… DNA MATCH.]” “Oh, my God. It's us.”

I want this preserved for posterity, because this made me absolutely hoot. They avoided fake science technobabble by going so far in the other direction that it becomes equally meaningless.
What the scene is trying to say is “this alien shares the vast majority of its genetic material with humans, indicating that they are in fact related.”
I will get to how one would actually determine that, but first: The head turned into green goop. Green goop. Humans are notably not prone to turning into green goop. Otherwise Nickelodeon would’ve probably been shut down within a week.

(Image credit: Rich Fury/KCA2021/Getty Images for Nickelodeon)
This annoyed me so much that years later, I dug up a possible explanation that backfills this with cool biology.
Humans, and almost all vertebrates have hemoglobin-filled blood. And on a tangent that I must follow: The only vertebrate that doesn’t is the icefish Channichthyidae family, commonly known as the white-blooded fish.
You’ll never guess what’s special about them.

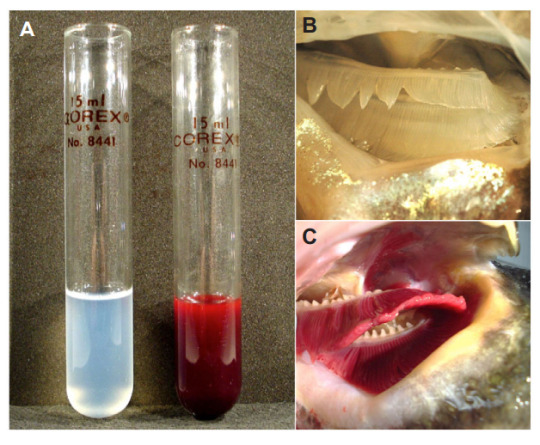
Yes, somehow these fish manage to live without hemoglobin, their blood only having 10% of the oxygen carrying capacity of their red-blooded cousins. Hell, most of them also lack myoglobin, which stores oxygen in muscle. The loss of myoglobin isn’t just a one-off event either, genetic studies have shown that these icefish have seen four distinct branches of their family tree lose myoglobin independently of each other. They have a wild series of adaptations to permit this, but basically they were already in such cold oxygen-rich water and moving so slow that they didn’t need all that extra oxygen-having stuff. They lost it, kept going, got bigger hearts, weirder muscles, and just. Kept. Going. They’ve actually expanded their range in the past 30 million years or so!
I love them! Evolution is wild. You know what’s also wild? There’s green-blooded vertebrates. Yes. You read that right. Yes, they still have hemoglobin. What they also have are staggering levels of biliverdin, which human bodies only produce when breaking down hemoglobin–when a bruise takes on a greenish hue, it’s because the dismantling of the blood under your skin has created biliverdin. While it’s generally been thought of as just a breakdown product, some research suggests that it also has protective effects against a number of diseases. In moderation, though. If you have enough of it to actually turn a bit green, you’ve got jaundice, which is not a thing you want to have.
But for a number of fish species, bush frogs, and skinks, they have way more biliverdin.
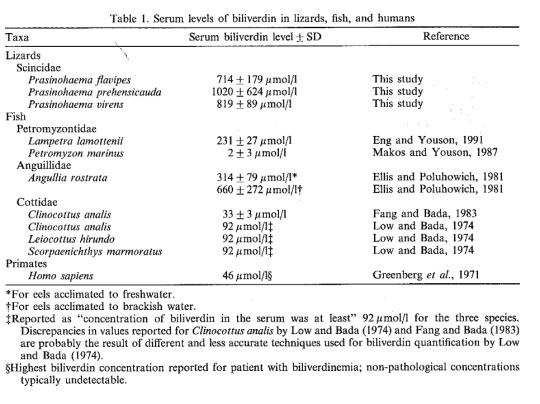
(Austin, C. C., & Jessing, K. W. (1994). Green-blood pigmentation in lizards. Comparative Biochemistry and Physiology Part A: Physiology, 109(3), 619-626.)
Humans usually don’t have much circulating biliverdin at all, so the table above compares someone with untreated jaundice to a number of other species–fish with two to fourteen times that amount, and the green-blooded skinks have twenty-two times as much! These creatures have green blood and turquoise-colored bones, and we still don’t know why. Maybe it’s protection against diseases, maybe it’s protection against parasites like malaria, maybe it’s to make them really blend in with foliage. Could be all of those at once, could be none of them, we don’t know! What we do know is that, as with the icefish, the green-blooded skinks in particular have independently evolved this feature four different times. (Rodriguez, Z. B., Perkins, S. L., & Austin, C. C. (2018). Multiple origins of green blood in New Guinea lizards. Science Advances, 4(5), eaao5017.)

(https://australian.museum/blog-archive/amri-news/amri-three-tiny-green-blooded-frogs-sing-like-birds/)

(https://web.archive.org/web/20180619143048/https://blog.nationalgeographic.org/2013/09/30/why-do-mysterious-lizards-have-green-blood/)
We have no mammals identified with biliverdin-filled green blood, you would need a lot of tweaks to how our bodies function to make this work. But it’s not literally impossible, like I thought in the theater! I’m quite sure the prop department didn’t do this level of research on the subject, but think about it!
I love biology! It’s! So! Weird!
And because I love biology, you’re not getting rid of me yet. My chosen field is genetics. This movie has presented me with a laughable sci-fi depiction of what we do.
So! What do we actually do, when we want to find out how related we are to another species?
I’m going to get into excruciating detail, so here’s the top-line summary: We extract the DNA, mash it up into readable little chunks, use some wicked cool machines to do the actual reading, and then we compare the target DNA with our DNA, and do some cooler stuff the movie isn’t aware of. A competent analysis would not only be able to tell you how much overlap two genomes have, but also be able to estimate how long the two species have been genetically distinct.
Is this way more than the movie needed for this plot point? Yes. But they didn’t actually have to do this at all, they could’ve just said the truth that science fiction usually ignores for budgetary reasons: “there’s no way these beings independently evolved to look so much like us, we have to be related.”
(Although even Star Trek, despite being the classic example of “putting a rubber thing on an actor’s forehead to make them an alien”, actually does acknowledge this. Precisely once. TNG s6e20, “The Chase”. It has never been mentioned again in the main line series, possibly because Rick Berman didn’t like it.)
Now. Time for me to take you all on a grand tour of DNA sequencing and phylogenetic analysis. You are all getting into the Willy Wonka boat with me. You have no choice.

So! You have a sample you’ve taken from a non-human mammal, one that’s never been genetically analyzed before. You are very lucky. You get to do fun stuff.

But before you get to sequencing, you have to purify any DNA in the sample. Your sample is full of all sorts of other biochemical gunk, and when cells are happy, DNA is packed away in the nucleus–you need to crack those open to get at the DNA.
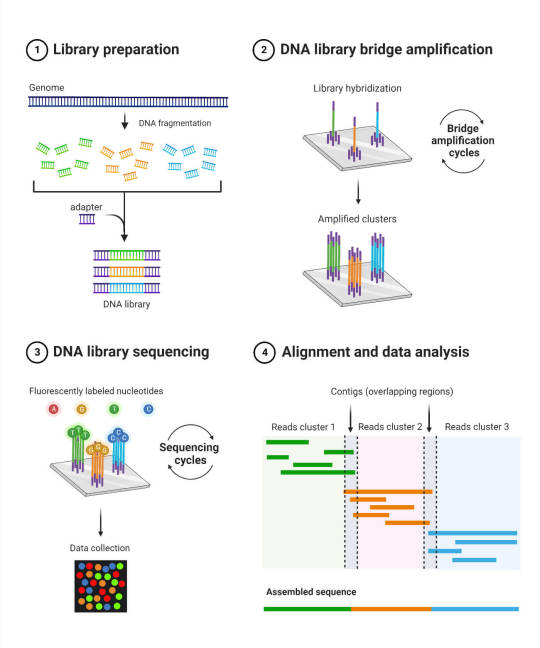
Next, you need to break the DNA into chunks, that’s #1 on the diagram above. For most of the past twenty years, this has meant chunking DNA down into pieces 25-50 letters long–just enough to probably get something unique over most of the genome, though you will have some areas that look identical at that tiny scale. In recent times, we’ve been getting better and better at what’s called “long read sequencing”, which at this point means fragments of several thousand DNA letters in length–though that’s still pretty short, compared to human chromosomes though: the average length of a human chromosome is 134 million letters long.
Depending on the sequencing technology and its needs, the sample may also need “amplification”: getting copied over and over using a protein originally harvested from hotspring-loving bacteria (#2). I always love that bit just as a concept: it’s one of many places where the modern study of genetics uses the microscopic, biological machinery of proteins for our own use!
After everything’s prepared, Then the sequencing itself can occur. That too is wild–the most common versions these days use tiny little fluorescent proteins to tag each letter of the DNA and read the sequence of lights (#3!). Some use infinitesimally tiny electrical modulations as DNA passes by a microscopic reader. There’s loads of different ways, anything works, so long as it can be read by a computer.

All this takes place in machines that are either small enough to fit on a countertop, or big enough to look like a fridge, and come in Apple White or Cheap Plastic Appliance.


Because you have a new species, you’re building what we call a reference genome. This tries to capture as much of the entire genome sequence as possible. Here’s an interesting wrinkle, though–A lot of samples won’t be just DNA from your target species! You might be picking up microbial DNA along the way as well. That can be really interesting and worth knowing about, though! Some people spend their whole careers studying the genomes of microbes found on people’s skin, or in their bodies. You’ll be computationally sorting out which sequences are in contiguous, mammalian chromosomes, which are from mitochondrial DNA (those cute little powerhouses have their own genomes!), and which come from microbes.
At the end of it, you have sequenced an entire genome. Because you want to find out how related it is to humans, you compare it to our reference genome–The human reference genomes we use is an assembly made from multiple individuals.* We use the reference genome as a common point of comparison that we refer to when studying genetic variation.
*Though if you’re working with data form the Genome Reference Consortium as is usually standard, one anonymous African-European donor, RP11, is still the backbone of the reference, accounting for 70% of the latest assembly.

(https://mk.bcgsc.ca/telomere-to-telomere-human-genome-assembly/posters.mhtml)
So, we’d compare this new mammalian genome to our own–how much overlap would we find? A lot. How you define our similarities and differences from other species can change the answer, but you’ll expect a lot of overlap. Some areas of the genome diverge faster than others, others are highly conserved–generally the more stable it is, the more important it is for our function.
Through many, many, many studies and corroboration with the archaeological record, we’ve worked out how to estimate how long ago two species diverged from each other. Actually, you’d rarely be comparing between just two species at this stage–get out all the other relevant reference genomes you’ve got! Compare them all! Build a phylogenetic tree–the modern version of that “tree of life” idea that Darwin popularized. Then you’ll have a more accurate sense of how your mystery species relates to everything.
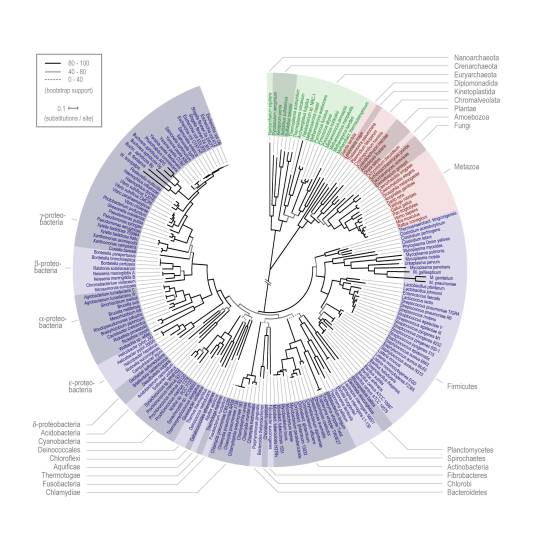
(https://www.embl.org/news/science/a-new-tree-of-life-allows-a-closer-look-at-the-origin-of-species/)
I’m going to go off on a tangent to end this post, because that’s just the start, taking the entire genome of a single individual. This is what most people think is what we always do. But no! That’s expensive overkill for most experiments. Once you’re familiar with a species, and you’ve sequenced DNA from many individuals, you can identify areas where lots of them have sequence variants. These can be completely benign, differences that make us all unique, or make an individual more susceptible to disease. This allows us to target what we want out of DNA sequencing: Are we trying to diagnose an illness? Identify a person from a tissue sample? Or are we doing something more exploratory?
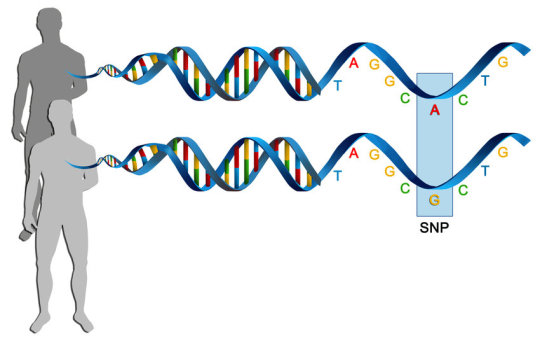
Depending on what you want, you select anywhere from a handful of locations, up to tens of thousands. The closest many people have gotten to this stuff is through ancestry services like 23andMe, which uses this kind of sequencing.
But that’s not all! There’s so many different targets to choose from, depending on what you want to do! So many techniques to get DNA in different ways! And we still haven’t gotten to the part that I actually work on.

I’m a computational geneticist. I get all the gigabytes of data that comes out of these experiments and I get to dig into the details, the patterns that emerge between genetic code and the details of the organisms we study, the connections between genome sequence and other, wilder things we can collect data on, the dizzying complexities of what goes on every microsecond of every day in every cell in your body.
I love my job.
⛬
(Previous) | (Index) | (Next)
⛬
Citations for alt-text rambles:
1. https://www.youtube.com/watch?v=O_YuTMDkWfI 2. https://doi.org/10.1242/jeb.116129 note: this source lists the image as credit to “J.M.B.”, which is not how I’m used to seeing images credited. Those are the initials of one of the authors, but I thought it meant “Journal of Molecular Biology”, so I went on a half hour wander around the internet trying to find where the hell this fish blood came from. 3. https://www.thebhs.org/publications/the-herpetological-journal/volume-13-number-4-october-2003/1729-01-hyperbiliverdinemia-in-the-shingleback-lizard-tiliqua-rugosa 4. https://doi.org/10.1111/j.1439-0264.2009.00952.x 5. https://en.wikipedia.org/wiki/Mandrill#Characteristics 6. https://www.vogue.com/article/dune-part-two-costumes-jacqueline-west-interview 7. http://dx.doi.org/10.13140/RG.2.2.29564.08327 8. https://www.researchgate.net/publication/357946568_New_approaches_and_concepts_to_study_complex_microbial_communities 9. https://karobben.github.io/2023/10/30/Bioinfor/PacBio/
Overflow Ramble 1
the fuckin “DNA MATCH” machine. I already wrote a 380-word alt-text about this thing last time. I’m not doing it again. I’m going to talk about things I like instead. Such as Dune: Part Two! Yes! I mentioned it last week and then didn’t ramble about it after seeing it. Well, NOW I WILL. tl;dr it’s good, go see it. I only vaguely remember the book, but I liked the changes they made to center the fact that no, Paul becoming Lisan al-Gaib is not actually a good thing.
Man, it’s nice to see a movie where the costumers and set designers got good time to work on their craft. (cite 6) Even the generic Harkonnen soldiers looked great–reminded me a lot of my beloved Warframe, probably because the costume designer was using H.R. Giger for inspiration there. Everything felt real. Even the stuff that definitely wasn’t–the gigantic spice harvesters and ships felt like living, physically present beings. The sand worms looked great. The movie did a fantastic job visually communicating the massive size of so many things. Especially because the camera remains restrained: no weightless zipping around, the camera itself follows paths and finds locations that make sense.
Chakobsa continues to be a fantastic conlang, now the work of both David and Jesse Peterson. It’s heard a lot more in this movie, and there are some great flourishes with it. While there isn’t as much Arabic vocabulary in it as in the original books, I remember from DJP’s work streams that he definitely was using the grammar of Arabic as one of his touchstones. Most key words remain Arabic though–jihad was removed, but it made me double-take in the theater when Stilgar referred to Paul as the Mahdi.
I’m of two minds about lowering the Arab influences on the Fremen–on the one hand, missing representation, which included some explicit ties to real world anti-imperial struggles in North Africa and the Middle East. On the other, these first two movies are about how the Fremen are manipulated by a colonial power, using their adherence to a faith that was manipulated by a different colonial power. They become both hapless victims and also perpetrators of colonial violence, with only Chani seeing through it.
I think the general decisionmaking process on cultural changes was motivated by a desire to remove some of Frank Herbert’s bad ideas–particularly around the Harkonnens, thank fuck. That seems to have been the thinking around altering the Fremen a bit as well. Did it succeed? Not my place to say. On all other notes, I have no reservations recommending the movie. It’s a very earnest attempt to bring that world to life, and I think it succeeds.
Overflow Ramble 2
A figure showing the basic steps of the standard Illumina sequencing method (cite 7). It is broken into four sub-figures:
Library Preparation. The genome is snipped into small fragments, then adapters are attached (“ligated”) to stabilize the molecule and make it behave. This creates a “library” of DNA that will be read from.
DNA library bridge amplification. The adapters on DNA fragments stick to a prepared plate, which is covered in little clusters of molecules that specially attach to those adapters. Biochemical processes are then carried out in repeated cycles to duplicate (or “amplify”) those fragments in such a way that the clusters on the plate are all filled with copies of just one DNA fragment.
DNA library sequencing. The DNA is modified so that the four letters it’s made out of all glow a specific color, with each DNA letter shining in sequence. This is pure awesome and I love it.
Alignment and data analysis. Because of some details on how step 1 is done, you have lots of fragments that create an overlapping patchwork of sequences. This allows (most of) the genome to be pasted back together by looking for overlaps (“contiguous sequences”, or “contigs” for short).
Congratulations! You have just attended an abridged graduate-level introductory lecture on Illumina sequencing.
Overflow Ramble 3
A diagram of PacBio Systems’ sequencing technology, Single Molecule, Real-Time Sequencing, or SMRT Sequencing, because scientists love acronyms. Pretty much every step is different from how Illumina does it. I cannot find a diagram that’s both brief and also good at explaining what it’s showing, so this is the best I could find. It’s split into four parts with attendant text, which I’ll try and explain as well.
“SMRTbell template. Two hairpin adapters allow continuous circular sequencing.”
Library preparation basically involves taking a longer chunk of DNA and splitting it in half lengthwise, in such a way that the two strands of DNA will form a single-stranded loop. This is called a SMRTbell library. Why? I have no idea!
“ZMW wells. Sites where sequencing takes place.”
Then, these are fed into SMRT Cells, which contain zero-mode waveguides (ZMWs). I was once told what this means, and I have completely forgotten, but it sounds like something from Gundam.
“Modified polymerase. As a nucleotide is incorporated by the polymerase, a camera records the emitted light.”
What I do understand is that at the bottom of each of these little holes, they stick a molecule which the DNA sticks to. This molecule, a polymerase, has precisely one job: make more DNA, an exact copy of what it’s latched onto. So you give it this loop of DNA, feed it a soup of free DNA letters, and it starts cranking out a new strand.
“PacBio output. A camera records the changing colours from all ZMWs; each colour change corresponds to one base.”
Each one of the DNA letters given to the polymerase has a special modifier, on it which flashes a color when the polymerase slots it into the new strand it’s making. A camera picks up this flash. And, because the DNA is a circle, the polymerase doesn’t know where to stop–it just keeps going and going until something breaks or it runs out of letters to work with. This means that even if the camera misses a flash the first time, it will have more chances to see it, and confirm what it already saw.
Wait what in the fuck this figure was from somebody’s thesis (cite 8), but that wasn’t what actually got this into search results. What got it there was a github page with a vtuber avatar sitting in the corner?? What??? (cite 9)
⛬
(Previous) | (Index) | (Next)
⛬
#Prometheus 2012#Prometheus (2012)#I go rogue and refuse to talk about the movie#or rather I spend 4600 words talking about a quarter of a scene as a springboard to talk about my job and how cool it is#I kind of envy people who work on wild population genetics#but also their sample sizes are tiny and bad for my particular specialty#so I'm giving a broad overview on how reference genome assembly works and I've only ever done phylogenetics on intraspecific variants#yes this is how much I talk about bits of my job that aren't even directly my job#because working in genetics is SO COOL#tomorrow we'll be back to analyzing inexplicable characters and androids poisoning people don't worry
45 notes
·
View notes