#biomolecularinteractions
Explore tagged Tumblr posts
Text
Explore Drugs Targeting RNA Riboswitches with Magna - Depixus
We’ve been working with Dr Jay Schneekloth, a leading expert in RNA-targeted drug discovery, to use our MAGNA™ technology to probe the interactions between the PreQ1 bacterial riboswitch and its natural ligand or a synthetic small molecule.
In a paper published as a preprint on bioRxiv and submitted for publication in a high impact journal, we showed how MAGNA provided novel insights into the distinct mechanisms of action of different ligands upon the target RNA at single molecule resolution.
What is the PreQ1 bacterial riboswitch?
Riboswitches are structured sequences of RNA that are commonly found in the 5′ untranslated regions of bacterial mRNAs and help to control metabolic pathways. They work through direct binding of a ligand such as a metabolite to the riboswitch, which induces a conformation change to their secondary structure and alters gene expression.
The PreQ1 riboswitch regulates downstream gene expression for the biosynthesis of queuosine in response to a metabolite called PreQ1 (7-aminomethyl-7-deazaguanine). Upon binding of PreQ1 the RNA changes into a shape known as a pseudoknot, which alters gene expression at either the transcriptional or translational level.
The ability of riboswitches to modulate bacterial gene expression in response to small molecules such as metabolites makes them an attractive group of novel antibacterial drug targets.
Using MAGNA to explore RNA-ligand interactions
Based on magnetic force spectroscopy, MAGNA is the first technology for exploring dynamic molecular interactions in real time from up to thousands of individual molecules. Here, we used it to probe the interactions between the PreQ1 riboswitch RNA from Bacillus subtilis (Bsu) and its natural ligand, PreQ1, or a synthetic ligand named Compound 4.
MAGNA allows us to precisely measure the effects of applying constant or gradually changing (ramped) forces to hundreds of immobilized riboswitch RNAs simultaneously (Figure 1A). This enabled us to capture the exact point at which each RNA molecule changes conformation.
We found that both PreQ1 and Compound 4 stabilized the riboswitch RNA structure, requiring more force to unfold it (Figure 1B). We were also able to explore the concentration dependency of their effects (Figure 1C, E, F).
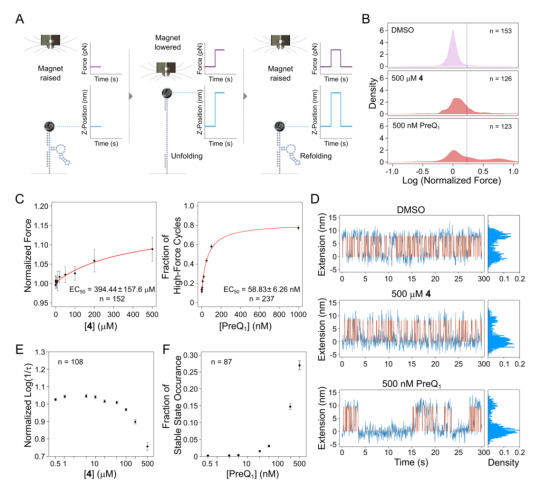
Figure 1: (A) Overview of MAGNA as a single-molecule platform for exploring the interactions of bioactive small molecule ligands with their target RNA structures in real-time (B) Unfolding force distributions of the Bsu PreQ1 riboswitch aptamer in control (DMSO), Compound 4 and PreQ1 ligand conditions (C) Dose-response curve for the change in unfolding of the aptamer in the presence of 4 and PreQ1 (D) Raw traces of the constant-force experiments for control, 4 and PreQ1 of a single molecule with cumulative density histograms shown to the right. (E) The aptamer unfolding rate as a function of the concentration of 4 in constant force experiments. (F) The impact of PreQ1 ligand concentration on the occurrence probability of the stable folded state. n is the number of molecules analyzed. Taken from Parmar et al (bioRxiv, 2024).
In addition, Schneekloth and colleagues explored other structural aspects of the interaction between the riboswitch and various ligands, and their impact on gene expression in live bacterial cells. They were also able to show that Compound 4 could bind in a similar way to PreQ1 riboswitches from other species of bacteria, building up a fuller picture of the properties of this novel molecule.
Deeper insights for RNA-targeted drug development
Overall, the findings presented in the paper demonstrate that even though different ligands may appear to be the same in terms of RNA binding site and impact on gene expression, other factors including selectivity, mode of recognition, and impacts on both conformational kinetics and thermodynamics, can all play
a role in the abili
ty of a compound to modulate biological function. These subtle yet important distinctions may not be apparent using analytical methods that rely on surrogate outputs or bulk measurements.
MAGNA’s unique single molecule approach enabled the Schneekloth team to visualize the binding of a novel ligand to its target in real time and gather insights into the distinct mechanisms of action, which would not have been possible using other methods.
With MAGNA we now have a technology that can not only identify compounds that bind to RNA, but also directly reveal how they impact its structure and function at the level of individual molecules. And we can use it to generate detailed information about the kinetics and concentration dependence of ligand binding.
These results demonstrate the capabilities of MAGNA to deliver valuable data about individual dynamic molecular interactions at scale, supporting lead selection in the development of novel RNA-targeted therapeutics.
Read More here: https://depixus.com/using-magna-to-explore-drugs-targeting-rna-riboswitches/
#TechnologyIntroduction#InnovativeTechnology#DepixusTechnology#MAGNATechnology#MAGNAbiomolecularinteractions#MAGNAdrugdiscovery#MAGNAproteinproteininteractions#Biomolecularinteractions#Moleculebinding#Drugdiscovery#Proteinproteininteractions#Biophysics#Structuralbiology#Moleculardynamics#Computationalbiology#RNA#Riboswitches#depixus
0 notes
Text
"Label-Free Detection: Enhancing Research Accuracy and Efficiency"
Label-free detection is a cutting-edge technology in the field of biochemical and pharmaceutical research, offering a sophisticated method to analyze biomolecular interactions without the need for fluorescent or radioactive labels. This technology provides real-time, high-throughput, and quantitative data, enhancing the accuracy and efficiency of drug discovery and development processes. Label-free detection methods, such as surface plasmon resonance (SPR), bio-layer interferometry (BLI), and optical biosensors, are revolutionizing how scientists study protein-protein, protein-DNA, and protein-small molecule interactions. The growing demand for more efficient and cost-effective research tools, coupled with the need to reduce interference from labels that can affect molecular behavior, is driving the adoption of label-free detection technologies. Moreover, advancements in instrumentation and data analysis software are making these technologies more accessible and user-friendly, further expanding their application in various research and diagnostic fields. Despite challenges like high initial costs and the need for specialized equipment, the future of label-free detection is promising, with ongoing innovations expected to enhance its capabilities and broaden its impact on scientific research and medical diagnostics.
#LabelFreeDetection #BiochemicalResearch #DrugDiscovery #SurfacePlasmonResonance #OpticalBiosensors #BioLayerInterferometry #BiomolecularInteractions #HealthTech #PharmaceuticalResearch #LabTech #ScientificInnovation #MolecularAnalysis #ResearchTools #Diagnostics #BioTech
0 notes
Text

Depixus Wins SLAS2024 Ignite Award for MAGNA Technology - Depixus
#biomolecularinteractions#singlemolecule#lifescience#biophysics#nanotechnology#SLAS2024#awardwinning#breakthroughtechnology#MAGNA#Depixus
0 notes
Text
Depixus Wins SLAS2024 Ignite Award for MAGNA Technology - Depixus

Paris, France – Interactomics pioneer Depixus has won the prestigious Society for Laboratory Automation and Screening (SLAS) Ignite Award at the society’s annual International Conference and Exhibition in Boston, MA, for their novel MAGNA™ technology. The award coincides with the first showing of Depixus’ ground-breaking MAGNA One™ instrument at the conference.
The Ignite Award recognizes exciting companies providing game-changing technologies for the life sciences. Judged by a panel of experts, participants were scored on excellence in innovation, marketing presence and opportunity, messaging, impact, funding, and balanced leadership.
Depixus was one of just 16 companies selected to participate in the SLAS2024 Innovation AveNEW program – a specially designated area for start-ups and emerging companies within the conference – of which the top eight were shortlisted as finalists for the Ignite Award.
Based on magnetic force spectroscopy, MAGNA One enables scalable analysis of dynamic biomolecular interactions. This technology is the first to offer direct, simultaneous, and real-time measurements from thousands of individual molecules.
It is particularly useful for exploring challenging targets such as RNA and protein-protein interactions, providing invaluable insights into disease mechanisms and accelerating the development of novel therapeutics.
Depixus CEO Gordon Hamilton says, “We’re thrilled to have won this year’s Ignite Award against such stiff competition and congratulate all the other finalists. This recognition is a great way to celebrate the company we are building, our innovative technology, and its potential to impact the understanding and treatment of diseases where new approaches are urgently needed.”Depixus Wins SLAS2024 Ignite Award for MAGNA Technology, Depixus Chief Commercial Officer Steve Klose added, “The timing of this award aligns perfectly with the launch of our technology access program and commercial rollout of MAGNA One. It’s been an honor to unveil our new instrument to the world at SLAS2024, and we’re excited to see how the scientific community puts it to work to transform research.”
#biomolecularinteractions#singlemolecule#lifescience#drugdiscovery#biophysics#nanotechnology#SLAS2024#awardwinning#breakthroughtechnology#MAGNA#Depixus
0 notes
Text
Depixus Wins SLAS2024 Ignite Award for MAGNA Technology
Depixus introduces MAGNA™, a groundbreaking technology that utilizes magnetic force spectroscopy to examine biomolecular interactions at the single-molecule level, unlocking a new era in disease research and therapeutic development. MAGNA™ recently won the prestigious SLAS2024 Ignite Award, a testament to its revolutionary potential.
#biomolecularinteractions#singlemolecule#lifescience#drugdiscovery#biophysics#nanotechnology#SLAS2024#awardwinning#breakthroughtechnology#MAGNA#Depixus
0 notes
Text
How MAGNA™ Technology is Revolutionizing the Study of Biomolecular Interactions
In the world of science, understanding how molecules interact with each other is crucial for developing new drugs, diagnostics, and other life-saving technologies. However, traditional methods for studying these interactions are often time-consuming, expensive, and inaccurate.
MAGNA™ technology is a new and innovative approach that is revolutionizing the way we study biomolecular interactions. This powerful tool can measure the strength, structure, and location of interactions between molecules with unprecedented precision and sensitivity.
What is MAGNA™ technology?
MAGNA™ technology is based on a novel method called bioluminescence resonance energy transfer (BRET), It is developed by Depixux. BRET uses light to measure the distance between two molecules. When two molecules that are labeled with special BRET tags come close together, they can transfer energy from one to the other, which produces a light signal. The intensity of the light signal is proportional to the distance between the molecules.
How does MAGNA™ technology work?
MAGNA™ technology can be used in three different modes:
Binding strength mode: In this mode, MAGNA™ is used to measure the force required to pull apart two molecules. This can be used to study the strength of drug-target interactions, protein-protein interactions, and other biomolecular interactions.
Binding structure mode: In this mode, MAGNA™ is used to measure the effect of another molecule on the structure of a target molecule. This can be used to study how drugs bind to their targets, how proteins interact with each other, and how other molecules can alter the structure of biomolecules.
Binding location mode: In this mode, MAGNA™ is used to measure the precise location on a molecule where another molecule binds. This can be used to study the binding sites of drugs, the active sites of enzymes, and other important regions of biomolecules.
What are the benefits of MAGNA™ technology?
MAGNA™ technology offers several advantages over traditional methods for studying biomolecular interactions:
High sensitivity: MAGNA™ can detect interactions between molecules that are too weak to be measured by other methods.
High precision: MAGNA™ can measure the distance between molecules with high accuracy.
Real-time: MAGNA™ can measure interactions in real time, which allows researchers to study the dynamics of biomolecular interactions.
Label-free: MAGNA™ does not require the molecules to be labeled with special tags, which can simplify experiments and reduce costs.
How is MAGNA™ technology being used?
MAGNA™ technology is being used in a variety of research applications, including:
Drug discovery: MAGNA™ is being used to identify new drug targets and to develop new drugs that are more effective and have fewer side effects.
Diagnostics: MAGNA™ is being used to develop new diagnostic tests for diseases such as cancer and Alzheimer's disease.
Protein engineering: MAGNA™ is being used to engineer proteins with new properties and functions.
The future of MAGNA™ technology
MAGNA™ technology is a powerful tool that is revolutionizing the way we study biomolecular interactions. This technology has the potential to lead to the development of new drugs, diagnostics, and other life-saving technologies. As MAGNA™ technology continues to develop by Depixus, we can expect to see even more exciting discoveries in the years to come.
#TechnologyIntroduction#InnovativeTechnology#DepixusTechnology#MAGNATechnology#MAGNAbiomolecularinteractions#MAGNAdrugdiscovery#MAGNAproteinproteininteractions#Biomolecularinteractions#Moleculebinding#Drugdiscovery#Proteinproteininteractions#Biophysics#Structuralbiology#Moleculardynamics#Computationalbiology#depixus
0 notes
Text
MAGNA™: Revolutionizing Biomolecular Interaction Measurement - Depixus
#TechnologyIntroduction#InnovativeTechnology#DepixusTechnology#MAGNATechnology#MAGNAbiomolecularinteractions#MAGNAdrugdiscovery#MAGNAproteinproteininteractions#Biomolecularinteractions#Moleculebinding#Drugdiscovery#Proteinproteininteractions#Biophysics#Structuralbiology#Moleculardynamics#Computationalbiology#depixus
0 notes